| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| host | mouse |
| isotype | IgG |
| clonality | monoclonal |
| concentration | concentrate, predilute |
| applications | IHC |
| reactivity | human |
| available size | 0.1 mL, 0.5 mL, 1 mL concentrated, 7 mL prediluted |
rabbit anti-IDH1 (R132H) monoclonal antibody (ZR7) 6220
Price range: $160.00 through $528.00
Antibody summary
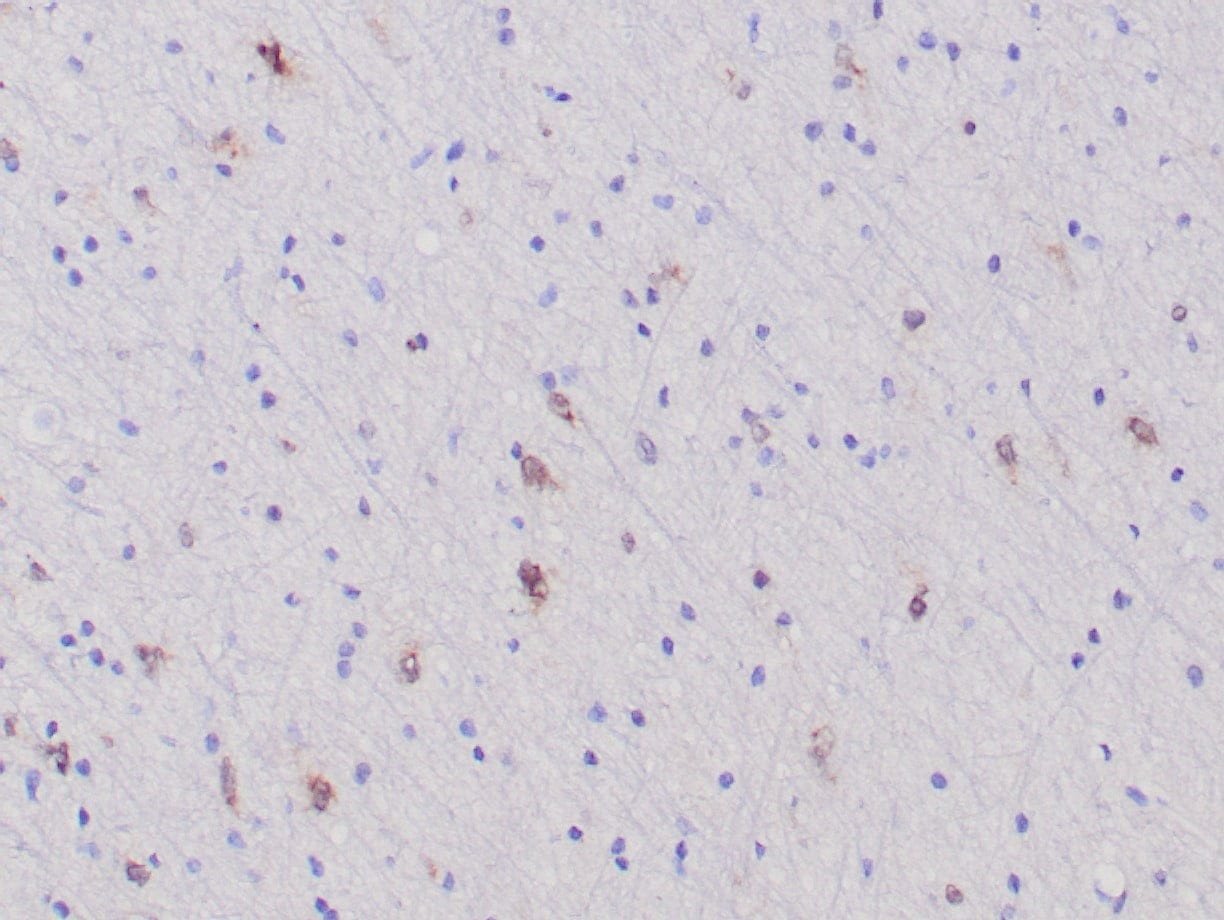
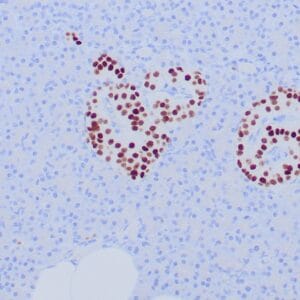
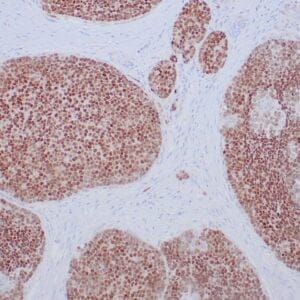
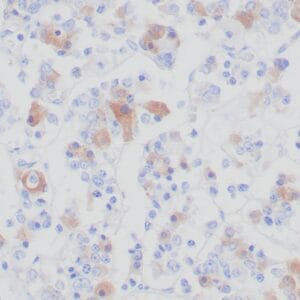
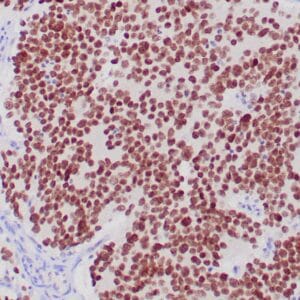
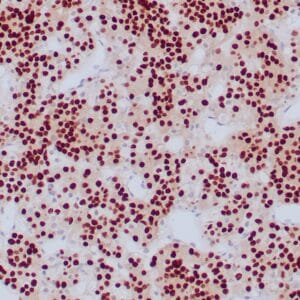
- Rabbit monoclonal to IDH1 (R132H)
- Suitable for: Immunohistochemistry (formalin-fixed, paraffin-embedded tissues)
- Reacts with: Human
- Isotype:IgG
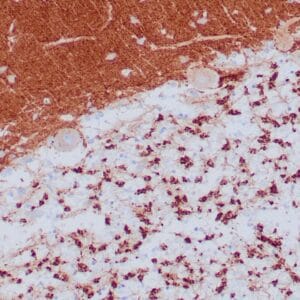
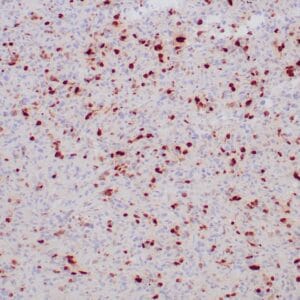
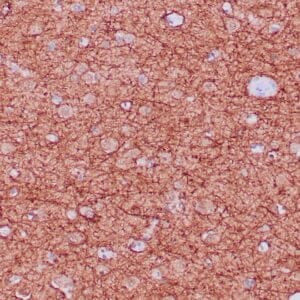
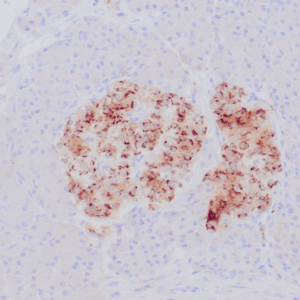
- Control: Infiltrating glioma
- Visualization: Nuclear
- 0.1, 0.5, 1.0 mL concentrated, 7 mL prediluted
rabbit anti-IDH1 (R132H) monoclonal antibody ZR7 6220
| target relevance |
|---|
| Protein names Isocitrate dehydrogenase [NADP] cytoplasmic (IDH) (IDH1) (EC 1.1.1.42) (Cytosolic NADP-isocitrate dehydrogenase) (IDPc) (NADP(+)-specific ICDH) (Oxalosuccinate decarboxylase) |
| Gene names IDH1,IDH1 PICD |
| Protein family Isocitrate and isopropylmalate dehydrogenases family |
| Mass 46659Da |
| Function FUNCTION: Catalyzes the NADP(+)-dependent oxidative decarboxylation of isocitrate (D-threo-isocitrate) to 2-ketoglutarate (2-oxoglutarate), which is required by other enzymes such as the phytanoyl-CoA dioxygenase (PubMed:10521434, PubMed:19935646). Plays a critical role in the generation of NADPH, an important cofactor in many biosynthesis pathways (PubMed:10521434). May act as a corneal epithelial crystallin and may be involved in maintaining corneal epithelial transparency (By similarity). {ECO:0000250|UniProtKB:Q9XSG3, ECO:0000269|PubMed:10521434, ECO:0000269|PubMed:19935646, ECO:0000303|PubMed:10521434}. |
| Catalytic activity CATALYTIC ACTIVITY: Reaction=D-threo-isocitrate + NADP(+) = 2-oxoglutarate + CO2 + NADPH; Xref=Rhea:RHEA:19629, ChEBI:CHEBI:15562, ChEBI:CHEBI:16526, ChEBI:CHEBI:16810, ChEBI:CHEBI:57783, ChEBI:CHEBI:58349; EC=1.1.1.42; Evidence={ECO:0000269|PubMed:10521434, ECO:0000269|PubMed:19935646}; PhysiologicalDirection=left-to-right; Xref=Rhea:RHEA:19630; Evidence={ECO:0000269|PubMed:19935646}; |
| Subellular location SUBCELLULAR LOCATION: Cytoplasm, cytosol {ECO:0000269|PubMed:10521434}. Peroxisome {ECO:0000269|PubMed:10521434}. |
| Structure SUBUNIT: Homodimer. {ECO:0000269|PubMed:15173171, ECO:0000269|PubMed:19935646}. |
| Post-translational modification PTM: Acetylation at Lys-374 dramatically reduces catalytic activity. {ECO:0000250}. |
| Involvement in disease DISEASE: Glioma (GLM) [MIM:137800]: Gliomas are benign or malignant central nervous system neoplasms derived from glial cells. They comprise astrocytomas and glioblastoma multiforme that are derived from astrocytes, oligodendrogliomas derived from oligodendrocytes and ependymomas derived from ependymocytes. {ECO:0000269|PubMed:19117336, ECO:0000269|PubMed:19935646}. Note=The gene represented in this entry is involved in disease pathogenesis. Mutations affecting Arg-132 are tissue-specific, and suggest that this residue plays a unique role in the development of high-grade gliomas. Mutations of Arg-132 to Cys, His, Leu or Ser abolish magnesium binding and abolish the conversion of isocitrate to alpha-ketoglutarate. Instead, alpha-ketoglutarate is converted to R(-)-2-hydroxyglutarate. Elevated levels of R(-)-2-hydroxyglutarate are correlated with an elevated risk of malignant brain tumors. {ECO:0000269|PubMed:19935646}.; DISEASE: Note=Genetic variations are associated with cartilaginous tumors such as enchondroma or chondrosarcoma. Mutations of Arg-132 to Cys, Gly or His abolish the conversion of isocitrate to alpha-ketoglutarate. Instead, alpha-ketoglutarate is converted to R(-)-2-hydroxyglutarate. {ECO:0000269|PubMed:26161668}. |
| Target Relevance information above includes information from UniProt accession: O75874 |
| The UniProt Consortium |
Data
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
| IHC |
Documents
| # | SDS | Certificate | |
|---|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | |||
Only logged in customers who have purchased this product may leave a review.


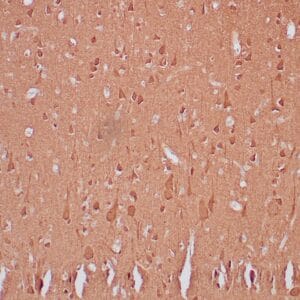
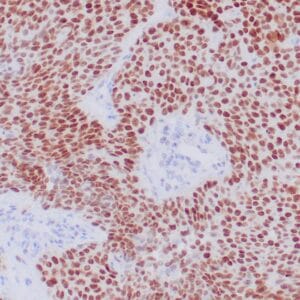
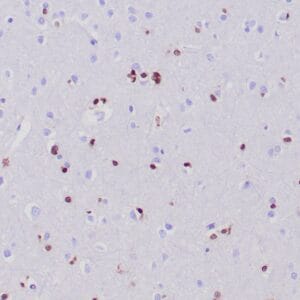
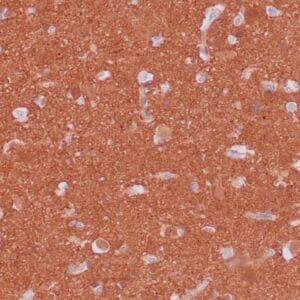
Reviews
There are no reviews yet.