| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| host | mouse |
| isotype | IgG |
| clonality | monoclonal |
| concentration | concentrate, predilute |
| applications | IHC |
| reactivity | human |
| available size | 0.1 mL, 0.5 mL, 1 mL concentrated, 7 mL prediluted |
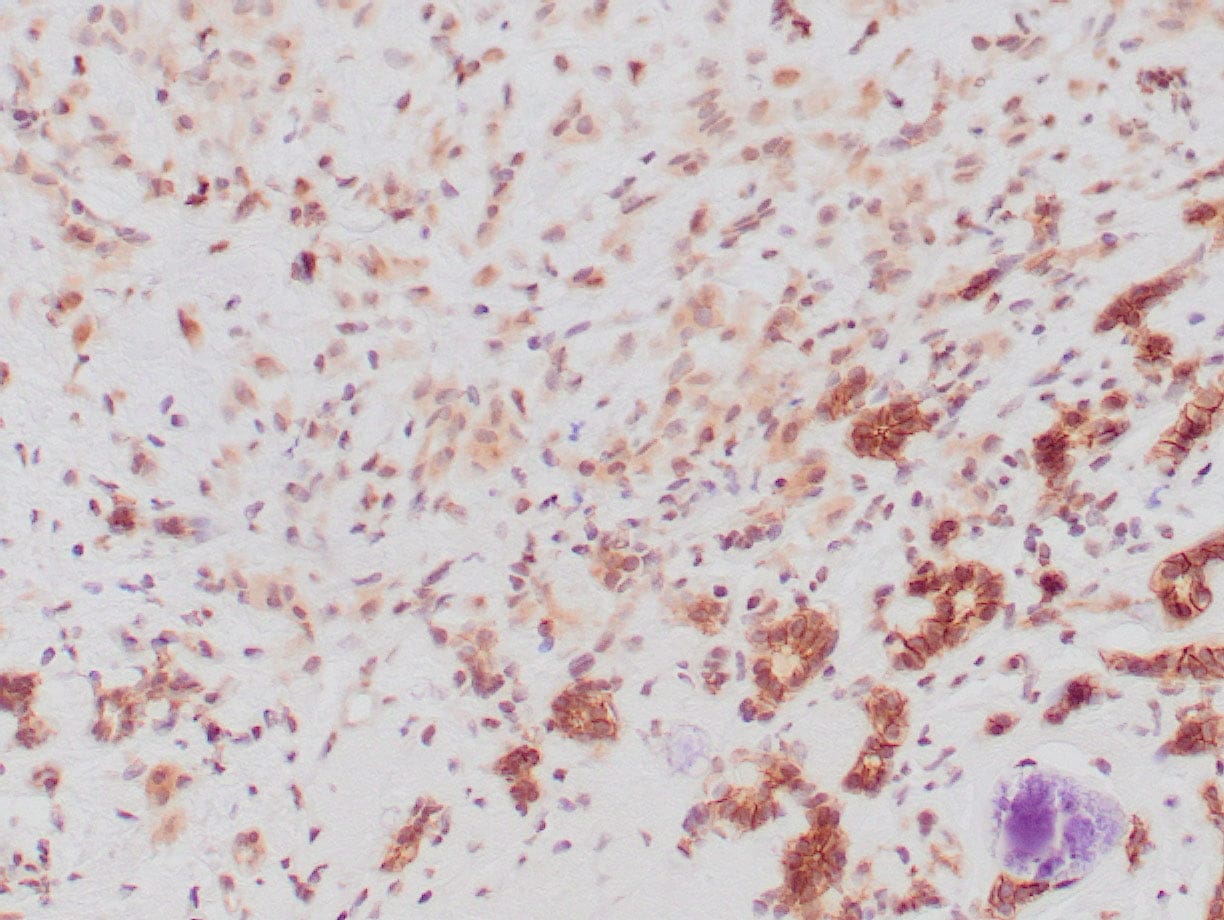
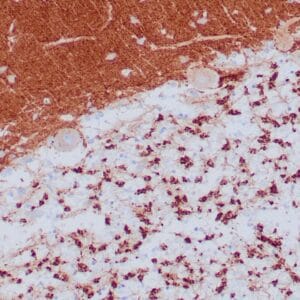
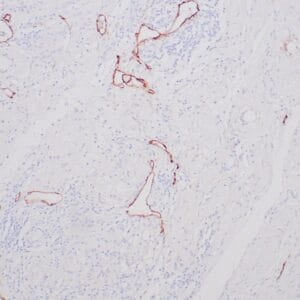
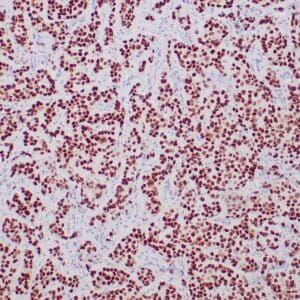
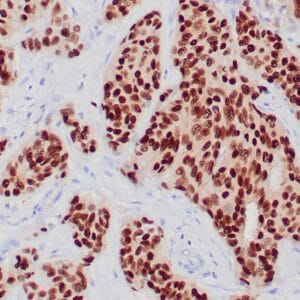
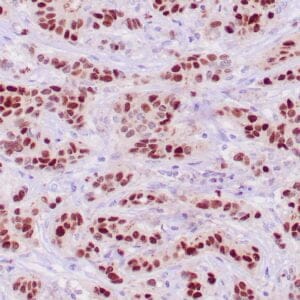
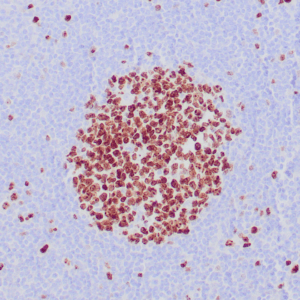
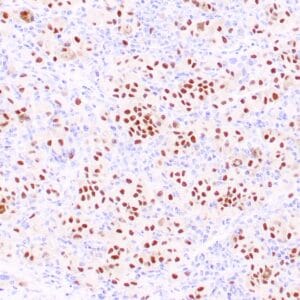
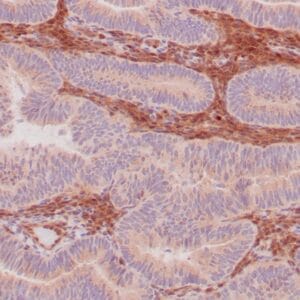
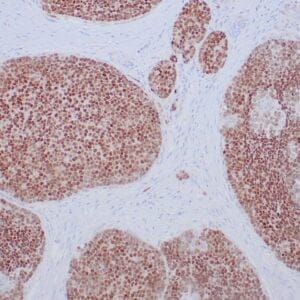
rabbit anti-p120 monoclonal antibody (ZR316) 6302
Price range: $160.00 through $528.00
Antibody summary
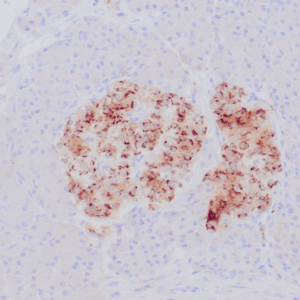
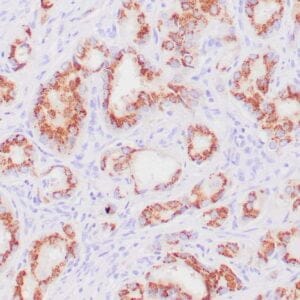
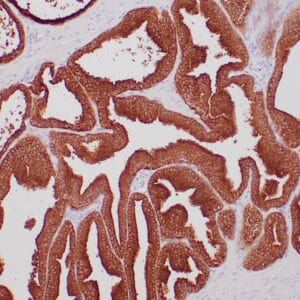
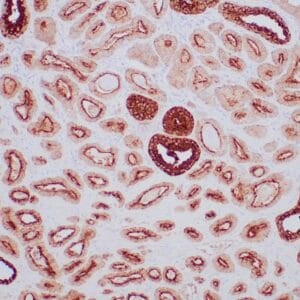
- Rabbit monoclonal to p120
- Suitable for: Immunohistochemistry (formalin-fixed, paraffin-embedded tissues)
- Reacts with: Human
- Isotype:IgG
- Control: Breast lobular carcinoma, melanoma
- Visualization: Cytoplasmic and membranous
- 0.1, 0.5, 1.0 mL concentrated, 7 mL prediluted
rabbit anti-p120 monoclonal antibody ZR316 6302
| target relevance |
|---|
| Protein names Catenin delta-1 (Cadherin-associated Src substrate) (CAS) (p120 catenin) (p120(ctn)) (p120(cas)) |
| Gene names CTNND1,CTNND1 KIAA0384 |
| Protein family Beta-catenin family |
| Mass 108170Da |
| Function FUNCTION: Key regulator of cell-cell adhesion that associates with and regulates the cell adhesion properties of both C-, E- and N-cadherins, being critical for their surface stability (PubMed:14610055, PubMed:20371349). Promotes localization and retention of DSG3 at cell-cell junctions, via its interaction with DSG3 (PubMed:18343367). Beside cell-cell adhesion, regulates gene transcription through several transcription factors including ZBTB33/Kaiso2 and GLIS2, and the activity of Rho family GTPases and downstream cytoskeletal dynamics (PubMed:10207085, PubMed:20371349). Implicated both in cell transformation by SRC and in ligand-induced receptor signaling through the EGF, PDGF, CSF-1 and ERBB2 receptors (PubMed:17344476). {ECO:0000269|PubMed:10207085, ECO:0000269|PubMed:14610055, ECO:0000269|PubMed:17344476, ECO:0000269|PubMed:18343367, ECO:0000269|PubMed:20371349}. |
| Subellular location SUBCELLULAR LOCATION: Cell junction, adherens junction {ECO:0000269|PubMed:11896187}. Cytoplasm {ECO:0000269|PubMed:15240885, ECO:0000269|PubMed:17047063}. Nucleus {ECO:0000269|PubMed:11896187, ECO:0000269|PubMed:17115030}. Cell membrane {ECO:0000269|PubMed:15240885, ECO:0000269|PubMed:17047063}. Cell junction {ECO:0000269|PubMed:18343367}. Note=Interaction with GLIS2 promotes nuclear translocation (By similarity). Detected at cell-cell contacts (PubMed:15240885, PubMed:17047063). NANOS1 induces its translocation from sites of cell-cell contact to the cytoplasm (PubMed:17047063). CDH1 enhances cell membrane localization (PubMed:15240885). Localizes to cell-cell contacts as keratinocyte differentiation progresses (By similarity). {ECO:0000250|UniProtKB:P30999, ECO:0000269|PubMed:11896187, ECO:0000269|PubMed:15240885, ECO:0000269|PubMed:17047063}.; SUBCELLULAR LOCATION: [Isoform 1A]: Nucleus {ECO:0000269|PubMed:11896187}.; SUBCELLULAR LOCATION: [Isoform 2A]: Nucleus {ECO:0000269|PubMed:11896187}.; SUBCELLULAR LOCATION: [Isoform 3A]: Nucleus {ECO:0000269|PubMed:11896187}.; SUBCELLULAR LOCATION: [Isoform 4A]: Cytoplasm {ECO:0000269|PubMed:11896187}.; SUBCELLULAR LOCATION: [Isoform 1AB]: Cytoplasm {ECO:0000269|PubMed:11896187}. |
| Tissues TISSUE SPECIFICITY: Expressed in vascular endothelium. Melanocytes and melanoma cells primarily express the long isoform 1A, whereas keratinocytes express shorter isoforms, especially 3A. The shortest isoform 4A, is detected in normal keratinocytes and melanocytes, and generally lost from cells derived from squamous cell carcinomas or melanomas. The C-terminal alternatively spliced exon B is present in the p120ctn transcripts in the colon, intestine and prostate, but lost in several tumor tissues derived from these organs. {ECO:0000269|PubMed:11896187, ECO:0000269|PubMed:14699141}. |
| Structure SUBUNIT: Belongs to a multiprotein cell-cell adhesion complex that also contains E-cadherin/CDH1, alpha-catenin/CTNNA1, beta-catenin/CTNNB1, and gamma-catenin/JUP (PubMed:15240885, PubMed:20371349). Component of a cadherin:catenin adhesion complex composed of at least of CDH26, beta-catenin/CTNNB1, alpha-catenin/CTNNA1 and p120 catenin/CTNND1 (PubMed:28051089). Binds to the C-terminal fragment of PSEN1 and mutually competes for CDH1. Interacts with ZBTB33 (PubMed:10207085). Interacts with GLIS2 (PubMed:17344476). Interacts with FER (PubMed:7623846). Interacts with NANOS1 (via N-terminal region) (PubMed:17047063). Interacts (via N-terminus) with GNA12; the interaction regulates CDH1-mediated cell-cell adhesion (PubMed:15240885). Interacts with GNA13 (PubMed:15240885). Interacts with CCDC85B (PubMed:25009281). Interacts with PLPP3; negatively regulates the PLPP3-mediated stabilization of CTNNB1 (PubMed:20123964). Interacts with DSG3; the interaction facilitates DSG3 localization and retention at cell-cell junctions (PubMed:18343367). Interacts with CTNND1/p120-catenin; the interaction controls CADH5 endocytosis (By similarity). {ECO:0000250|UniProtKB:P30999, ECO:0000269|PubMed:10207085, ECO:0000269|PubMed:15240885, ECO:0000269|PubMed:17047063, ECO:0000269|PubMed:17344476, ECO:0000269|PubMed:18343367, ECO:0000269|PubMed:20123964, ECO:0000269|PubMed:20371349, ECO:0000269|PubMed:25009281, ECO:0000269|PubMed:28051089, ECO:0000269|PubMed:7623846}. |
| Post-translational modification PTM: Phosphorylated by FER and other protein-tyrosine kinases. Phosphorylated at Ser-288 by PAK5. Dephosphorylated by PTPRJ. {ECO:0000269|PubMed:12370829, ECO:0000269|PubMed:17194753, ECO:0000269|PubMed:20564219, ECO:0000269|PubMed:7623846}. |
| Domain DOMAIN: A possible nuclear localization signal exists in all isoforms where Asp-626--631-Arg are deleted.; DOMAIN: ARM repeats 1 to 5 mediate interaction with cadherins. |
| Involvement in disease DISEASE: Blepharocheilodontic syndrome 2 (BCDS2) [MIM:617681]: A form of blepharocheilodontic syndrome, a rare autosomal dominant disorder. It is characterized by lower eyelid ectropion, upper eyelid distichiasis, euryblepharon, bilateral cleft lip and palate, and features of ectodermal dysplasia, including hair anomalies, conical teeth and tooth agenesis. An additional rare manifestation is imperforate anus. There is considerable phenotypic variability among affected individuals. {ECO:0000269|PubMed:28301459}. Note=The disease is caused by variants affecting the gene represented in this entry. |
| Target Relevance information above includes information from UniProt accession: O60716 |
| The UniProt Consortium |
Data
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
| IHC |
Documents
| # | SDS | Certificate | |
|---|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | |||
Only logged in customers who have purchased this product may leave a review.


Reviews
There are no reviews yet.