| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| host | rabbit |
| isotype | IgG |
| clonality | monoclonal |
| concentration | 40 µg/mL |
| applications | IP, WB |
| reactivity | human, mouse |
| available sizes | 100 µL, 20 µL |
rabbit anti-FOXO1 polyclonal antibody blr140h 1024
Price range: $100.00 through $300.00
Antibody summary
- Rabbit polyclonal to FOXO (Forkhead box O) Transcription Factors
- Suitable for: IP,WB
- Reacts with: Hu, Ms
- Isotype: IgG
- 100 µL (10 blots), 20 µL
rabbit anti- foxo1 polyclonal antibody blr140h 1024
| target relevance |
|---|
| Protein names Forkhead box protein O1 (Forkhead box protein O1A) (Forkhead in rhabdomyosarcoma) |
| Gene names FOXO1,FOXO1 FKHR FOXO1A |
| Mass 69662Da |
| Function FUNCTION: Transcription factor that is the main target of insulin signaling and regulates metabolic homeostasis in response to oxidative stress (PubMed:10358076, PubMed:12228231, PubMed:15220471, PubMed:15890677, PubMed:18356527, PubMed:19221179, PubMed:20543840, PubMed:21245099). Binds to the insulin response element (IRE) with consensus sequence 5'-TT[G/A]TTTTG-3' and the related Daf-16 family binding element (DBE) with consensus sequence 5'-TT[G/A]TTTAC-3' (PubMed:10358076). Activity suppressed by insulin (PubMed:10358076). Main regulator of redox balance and osteoblast numbers and controls bone mass (By similarity). Orchestrates the endocrine function of the skeleton in regulating glucose metabolism (By similarity). Also acts as a key regulator of chondrogenic commitment of skeletal progenitor cells in response to lipid availability: when lipids levels are low, translocates to the nucleus and promotes expression of SOX9, which induces chondrogenic commitment and suppresses fatty acid oxidation (By similarity). Acts synergistically with ATF4 to suppress osteocalcin/BGLAP activity, increasing glucose levels and triggering glucose intolerance and insulin insensitivity (By similarity). Also suppresses the transcriptional activity of RUNX2, an upstream activator of osteocalcin/BGLAP (By similarity). Acts as an inhibitor of glucose sensing in pancreatic beta cells by acting as a transcription repressor and suppressing expression of PDX1 (By similarity). In hepatocytes, promotes gluconeogenesis by acting together with PPARGC1A and CEBPA to activate the expression of genes such as IGFBP1, G6PC1 and PCK1 (By similarity). Also promotes gluconeogenesis by directly promoting expression of PPARGC1A and G6PC1 (PubMed:17024043). Important regulator of cell death acting downstream of CDK1, PKB/AKT1 and STK4/MST1 (PubMed:18356527, PubMed:19221179). Promotes neural cell death (PubMed:18356527). Mediates insulin action on adipose tissue (By similarity). Regulates the expression of adipogenic genes such as PPARG during preadipocyte differentiation and, adipocyte size and adipose tissue-specific gene expression in response to excessive calorie intake (By similarity). Regulates the transcriptional activity of GADD45A and repair of nitric oxide-damaged DNA in beta-cells (By similarity). Required for the autophagic cell death induction in response to starvation or oxidative stress in a transcription-independent manner (PubMed:20543840). Mediates the function of MLIP in cardiomyocytes hypertrophy and cardiac remodeling (By similarity). Positive regulator of apoptosis in cardiac smooth muscle cells as a result of its transcriptional activation of pro-apoptotic genes (PubMed:19483080). Regulates endothelial cell (EC) viability and apoptosis in a PPIA/CYPA-dependent manner via transcription of CCL2 and BCL2L11 which are involved in EC chemotaxis and apoptosis (PubMed:31063815). {ECO:0000250|UniProtKB:A4L7N3, ECO:0000250|UniProtKB:G3V7R4, ECO:0000250|UniProtKB:Q9R1E0, ECO:0000269|PubMed:10358076, ECO:0000269|PubMed:12228231, ECO:0000269|PubMed:15220471, ECO:0000269|PubMed:15890677, ECO:0000269|PubMed:17024043, ECO:0000269|PubMed:18356527, ECO:0000269|PubMed:19221179, ECO:0000269|PubMed:19483080, ECO:0000269|PubMed:20543840, ECO:0000269|PubMed:21245099, ECO:0000269|PubMed:31063815}. |
| Subellular location SUBCELLULAR LOCATION: Cytoplasm {ECO:0000269|PubMed:11237865, ECO:0000269|PubMed:11311120, ECO:0000269|PubMed:12228231, ECO:0000269|PubMed:19221179, ECO:0000269|PubMed:20543840, ECO:0000269|PubMed:21245099, ECO:0000269|PubMed:25009184, ECO:0000269|PubMed:31063815}. Nucleus {ECO:0000269|PubMed:11311120, ECO:0000269|PubMed:12228231, ECO:0000269|PubMed:20543840, ECO:0000269|PubMed:25009184, ECO:0000269|PubMed:31063815}. Note=Shuttles between the cytoplasm and nucleus. Largely nuclear in unstimulated cells (PubMed:11311120, PubMed:12228231, PubMed:19221179, PubMed:20543840, PubMed:21245099, PubMed:25009184). In osteoblasts, colocalizes with ATF4 and RUNX2 in the nucleus (By similarity). Serum deprivation increases localization to the nucleus, leading to activate expression of SOX9 and subsequent chondrogenesis (By similarity). Insulin-induced phosphorylation at Ser-256 by PKB/AKT1 leads, via stimulation of Thr-24 phosphorylation, to binding of 14-3-3 proteins and nuclear export to the cytoplasm where it is degraded by the ubiquitin-proteasomal pathway (PubMed:11237865, PubMed:12228231). Phosphorylation at Ser-249 by CDK1 disrupts binding of 14-3-3 proteins and promotes nuclear accumulation (PubMed:18356527). Phosphorylation by NLK results in nuclear export (By similarity). Translocates to the nucleus upon oxidative stress-induced phosphorylation at Ser-212 by STK4/MST1 (PubMed:19221179, PubMed:21245099). SGK1-mediated phosphorylation also results in nuclear translocation (By similarity). Retained in the nucleus under stress stimuli including oxidative stress, nutrient deprivation or nitric oxide (By similarity). Retained in the nucleus on methylation (By similarity). PPIA/CYPA stimulates its nuclear accumulation (PubMed:31063815). Deacetylation by SIRT6, promotes its translocation into the cytoplasm (PubMed:25009184). {ECO:0000250|UniProtKB:Q9R1E0, ECO:0000269|PubMed:11237865, ECO:0000269|PubMed:11311120, ECO:0000269|PubMed:12228231, ECO:0000269|PubMed:18356527, ECO:0000269|PubMed:19221179, ECO:0000269|PubMed:20543840, ECO:0000269|PubMed:21245099, ECO:0000269|PubMed:25009184, ECO:0000269|PubMed:31063815}. |
| Tissues TISSUE SPECIFICITY: Expressed in umbilical endothelial cells (at protein level) (PubMed:19483080). Abundantly expressed in skeletal muscle and ovary, with lower expression in the heart, placenta, lung, liver, pancreas, spleen, testis and small intestine (PubMed:9479491). Weakly expressed in the brain, thymus, prostate and mucosal lining of the colon (PubMed:9479491). {ECO:0000269|PubMed:19483080, ECO:0000269|PubMed:9479491}. |
| Structure SUBUNIT: Interacts with LRPPRC. Interacts with RUNX2; the interaction inhibits RUNX2 transcriptional activity and mediates the IGF1/insulin-dependent BGLAP expression in osteoblasts Interacts with PPP2R1A; the interaction regulates the dephosphorylation of FOXO1 at Thr-24 and Ser-256 leading to its nuclear import. Interacts (acetylated form) with PPARG. Interacts with XBP1 isoform 2; this interaction is direct and leads to FOXO1 ubiquitination and degradation via the proteasome pathway (By similarity). Interacts with NLK. Interacts with SIRT1; the interaction results in the deacetylation of FOXO1 leading to activation of FOXO1-mediated transcription of genes involved in DNA repair and stress resistance. Binds to CDK1. Interacts with the 14-3-3 proteins, YWHAG and YWHAZ; the interactions require insulin-stimulated phosphorylation on Thr-24, promote nuclear exit and loss of transcriptional activity. Interacts with SKP2; the interaction ubiquitinates FOXO1 leading to its proteasomal degradation. The interaction requires the presence of KRIT1. Interacts (via the C-terminal half) with ATF4 (via its DNA-binding domain); the interaction occurs in osteoblasts, regulates glucose homeostasis via suppression of beta-cell proliferation and subsequent decrease in insulin production. Interacts with PRMT1; the interaction methylates FOXO1, prevents PKB/AKT1 phosphorylation and retains FOXO1 in the nucleus. Interacts with EP300 and CREBBP; the interactions acetylate FOXO1. Interacts with SIRT2; the interaction is disrupted in response to oxidative stress or serum deprivation, leading to increased level of acetylated FOXO1, which promotes stress-induced autophagy by stimulating E1-like activating enzyme ATG7. Interacts (acetylated form) with ATG7; the interaction is increased in response to oxidative stress or serum deprivation and promotes the autophagic process leading to cell death. Interacts (via the Fork-head domain) with CEBPA; the interaction increases when FOXO1 is deacetylated. Interacts with WDFY2. Forms a complex with WDFY2 and AKT1 (By similarity). Interacts with CRY1 (By similarity). Interacts with PPIA/CYPA; the interaction promotes FOXO1 dephosphorylation, nuclear accumulation and transcriptional activity (PubMed:31063815). Interacts with TOX4; FOXO1 is required for full induction of TOX4-dependent activity and the interaction is inhibited by insulin (By similarity). Interacts (when phosphorylated on Ser-256) with STUB1/CHIP (PubMed:19483080). {ECO:0000250|UniProtKB:Q9R1E0, ECO:0000269|PubMed:11237865, ECO:0000269|PubMed:15220471, ECO:0000269|PubMed:15668399, ECO:0000269|PubMed:15890677, ECO:0000269|PubMed:18356527, ECO:0000269|PubMed:18951090, ECO:0000269|PubMed:19483080, ECO:0000269|PubMed:20543840, ECO:0000269|PubMed:31063815}. |
| Post-translational modification PTM: Phosphorylation by NLK promotes nuclear export and inhibits the transcriptional activity. In response to growth factors, phosphorylation on Thr-24, Ser-256 and Ser-322 by PKB/AKT1 promotes nuclear export and inactivation of transactivational activity. Phosphorylation on Thr-24 is required for binding 14-3-3 proteins. Phosphorylation of Ser-256 decreases DNA-binding activity and promotes the phosphorylation of Thr-24 and Ser-319, permitting phosphorylation of Ser-322 and Ser-325, probably by CDK1, leading to nuclear exclusion and loss of function. Stress signals, such as response to oxygen or nitric oxide, attenuate the PKB/AKT1-mediated phosphorylation leading to nuclear retention. Phosphorylation of Ser-329 is independent of IGF1 and leads to reduced function. Dephosphorylated on Thr-24 and Ser-256 by PP2A in beta-cells under oxidative stress leading to nuclear retention (By similarity). Phosphorylation of Ser-249 by CDK1 disrupts binding of 14-3-3 proteins leading to nuclear accumulation and has no effect on DNA-binding nor transcriptional activity. Phosphorylation by STK4/MST1 on Ser-212, upon oxidative stress, inhibits binding to 14-3-3 proteins and nuclear export. PPIA/CYPA promotes its dephosphorylation on Ser-256 (PubMed:31063815). {ECO:0000250|UniProtKB:Q9R1E0, ECO:0000269|PubMed:10358075, ECO:0000269|PubMed:10358076, ECO:0000269|PubMed:11237865, ECO:0000269|PubMed:11311120, ECO:0000269|PubMed:11980723, ECO:0000269|PubMed:18356527, ECO:0000269|PubMed:18786403, ECO:0000269|PubMed:19221179, ECO:0000269|PubMed:21245099, ECO:0000269|PubMed:31063815}.; PTM: Ubiquitinated by SKP2 (PubMed:15668399). Ubiquitination leads to proteasomal degradation (PubMed:19483080). Ubiquitinated by STUB1/CHIP; when Ser-256 is phosphorylated (PubMed:19483080). {ECO:0000269|PubMed:15668399, ECO:0000269|PubMed:19483080}.; PTM: Methylation inhibits AKT1-mediated phosphorylation at Ser-256 and is increased by oxidative stress. {ECO:0000250|UniProtKB:Q9R1E0}.; PTM: Acetylated (PubMed:15220471, PubMed:15890677, PubMed:18786403, PubMed:20543840). Acetylation at Lys-262, Lys-265 and Lys-274 are necessary for autophagic cell death induction (PubMed:20543840). Deacetylated by SIRT2 in response to oxidative stress or serum deprivation, thereby negatively regulating FOXO1-mediated autophagic cell death (PubMed:20543840). Once in the nucleus, acetylated by CREBBP/EP300 (PubMed:15220471, PubMed:15890677, PubMed:18786403). Acetylation diminishes the interaction with target DNA and attenuates the transcriptional activity. It increases the phosphorylation at Ser-256 (PubMed:15220471, PubMed:15890677, PubMed:18786403). Deacetylation by SIRT1 results in reactivation of the transcriptional activity (PubMed:15220471, PubMed:15890677, PubMed:18786403). Oxidative stress by hydrogen peroxide treatment appears to promote deacetylation and uncoupling of insulin-induced phosphorylation (PubMed:15220471, PubMed:15890677, PubMed:18786403). By contrast, resveratrol acts independently of acetylation (PubMed:15220471, PubMed:15890677, PubMed:18786403). Acetylated at Lys-423, promoting its localization to the nucleus and transcription factor activity (PubMed:25009184). Deacetylation at Lys-423 by SIRT6, promotes its translocation into the cytoplasm, preventing its transcription factor activity (PubMed:25009184). Deacetylation and subsequent inhibition by SIRT6 has different effects depending on cell types: it inhibits gluconeogenesis in hepatocytes, promotes glucose sensing in pancreatic beta-cells and regulates lipid catabolism in brown adipocytes (By similarity). {ECO:0000250|UniProtKB:Q9R1E0, ECO:0000269|PubMed:15220471, ECO:0000269|PubMed:15890677, ECO:0000269|PubMed:18786403, ECO:0000269|PubMed:20543840, ECO:0000269|PubMed:25009184}. |
| Involvement in disease DISEASE: Rhabdomyosarcoma 2 (RMS2) [MIM:268220]: A form of rhabdomyosarcoma, a highly malignant tumor of striated muscle derived from primitive mesenchymal cells and exhibiting differentiation along rhabdomyoblastic lines. Rhabdomyosarcoma is one of the most frequently occurring soft tissue sarcomas and the most common in children. It occurs in four forms: alveolar, pleomorphic, embryonal and botryoidal rhabdomyosarcomas. Note=The gene represented in this entry may be involved in disease pathogenesis. Chromosomal aberrations involving FOXO1 are found in rhabdomyosarcoma. Translocation (2;13)(q35;q14) with PAX3 and translocation t(1;13)(p36;q14) with PAX7. The resulting protein is a transcriptional activator. {ECO:0000269|PubMed:8275086}. |
| Target Relevance information above includes information from UniProt accession: Q12778 |
| The UniProt Consortium |
Data
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
| Western blot |
Documents
| # | SDS | Certificate | |
|---|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | |||
Only logged in customers who have purchased this product may leave a review.



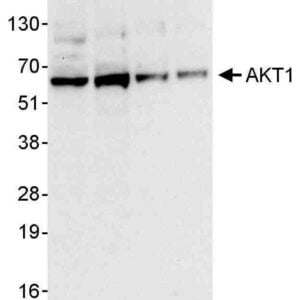
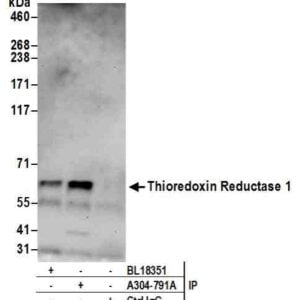
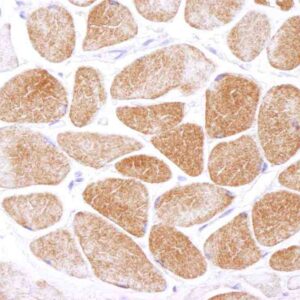
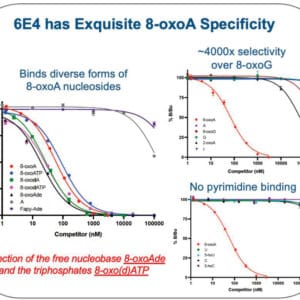

Reviews
There are no reviews yet.