| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| host | rabbit |
| isotype | IgG |
| clonality | polyclonal |
| concentration | 1 mg/mL |
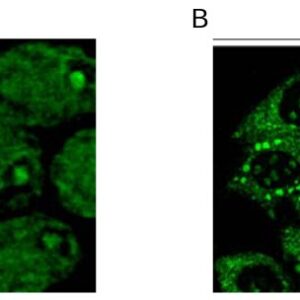
| applications | ICC/IF, WB |
| reactivity | Ezrin (Phospho-Tyr353) |
| available sizes | 100 µL |
rabbit anti-Ezrin (Phospho-Tyr353) polyclonal antibody 1249
$366.00
Antibody summary
- Rabbit polyclonal to Ezrin (Phospho-Tyr353)
- Suitable for: WB,IHC
- Isotype: Whole IgG
- 100 µl
rabbit anti-Ezrin (Phospho-Tyr353) polyclonal antibody 1249
| antibody |
|---|
| Tested applications WB,IHC,IHC |
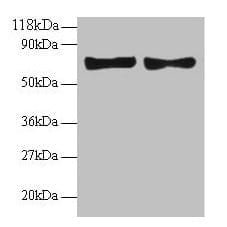
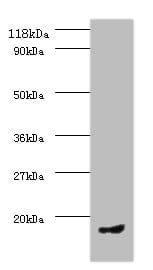
| Recommended dilutions Immunoblotting: use at dilution of 1:500-1:1,000. A band of ~81kDa is detected. Immunohistochemistry: use at dilution of 1:50- 1:100. These are recommended working dilutions. End user should determine optimal dilutions for their application. |
| Immunogen Peptide sequence that includes phosphorylation site of tyrosine 353 (Q-D-Y(p)-E-E) derived from human Ezrin and conjugated to KLH. |
| Size and concentration 100µL and 1 mg/mL |
| Form liquid |
| Storage Instructions This antibody is stable for at least one (1) year at -20°C. |
| Storage buffer PBS (without Mg2 and Ca2 ), pH 7.4, 150mM NaCl, |
| Purity affinity purified |
| Clonality polyclonal |
| Isotype IgG |
| Compatible secondaries goat anti-rabbit IgG, H&L chain specific, peroxidase conjugated, conjugated polyclonal antibody 9512 goat anti-rabbit IgG, H&L chain specific, biotin conjugated polyclonal antibody 2079 goat anti-rabbit IgG, H&L chain specific, FITC conjugated polyclonal antibody 7863 goat anti-rabbit IgG, H&L chain specific, Cross Absorbed polyclonal antibody 2371 goat anti-rabbit IgG, H&L chain specific, biotin conjugated polyclonal antibody, crossabsorbed 1715 goat anti-rabbit IgG, H&L chain specific, FITC conjugated polyclonal antibody, crossabsorbed 1720 |
| Isotype control Rabbit polyclonal - Isotype Control |
| target relevance |
|---|
| Protein names Ezrin (Cytovillin) (Villin-2) (p81) |
| Gene names EZR,EZR VIL2 |
| Mass 69413Da |
| Function FUNCTION: Probably involved in connections of major cytoskeletal structures to the plasma membrane. In epithelial cells, required for the formation of microvilli and membrane ruffles on the apical pole. Along with PLEKHG6, required for normal macropinocytosis. {ECO:0000269|PubMed:17881735, ECO:0000269|PubMed:18270268, ECO:0000269|PubMed:19111582}. |
| Subellular location SUBCELLULAR LOCATION: Apical cell membrane {ECO:0000269|PubMed:18046454}; Peripheral membrane protein {ECO:0000269|PubMed:18046454}; Cytoplasmic side {ECO:0000269|PubMed:18046454}. Cell projection {ECO:0000269|PubMed:18046454}. Cell projection, microvillus membrane {ECO:0000269|PubMed:18046454}; Peripheral membrane protein {ECO:0000269|PubMed:18046454}; Cytoplasmic side {ECO:0000269|PubMed:18046454}. Cell projection, ruffle membrane {ECO:0000269|PubMed:18046454}; Peripheral membrane protein {ECO:0000269|PubMed:18046454}; Cytoplasmic side {ECO:0000269|PubMed:18046454}. Cytoplasm, cell cortex {ECO:0000269|PubMed:18046454}. Cytoplasm, cytoskeleton {ECO:0000269|PubMed:18046454}. Cell projection, microvillus {ECO:0000250|UniProtKB:P26040}. Note=Localization to the apical membrane of parietal cells depends on the interaction with PALS1. Localizes to cell extensions and peripheral processes of astrocytes (By similarity). Microvillar peripheral membrane protein (cytoplasmic side). {ECO:0000250|UniProtKB:P31977}. |
| Tissues TISSUE SPECIFICITY: Expressed in cerebral cortex, basal ganglia, hippocampus, hypophysis, and optic nerve. Weakly expressed in brain stem and diencephalon. Stronger expression was detected in gray matter of frontal lobe compared to white matter (at protein level). Component of the microvilli of intestinal epithelial cells. Preferentially expressed in astrocytes of hippocampus, frontal cortex, thalamus, parahippocampal cortex, amygdala, insula, and corpus callosum. Not detected in neurons in most tissues studied. {ECO:0000269|PubMed:15797715}. |
| Structure SUBUNIT: Monomer. Homodimer (PubMed:27364155). Interacts with PALS1 and NHERF2. Found in a complex with EZR, PODXL and NHERF2 (By similarity). Interacts with MCC, PLEKHG6, PODXL, SCYL3/PACE1, NHERF1 and TMEM8B (PubMed:12651155, PubMed:15498789, PubMed:17616675, PubMed:17881735, PubMed:19555689, PubMed:9314537). Interacts (when phosphorylated) with FES/FPS (PubMed:18046454). Interacts with dimeric S100P, the interaction may be activating through unmasking of F-actin binding sites (PubMed:12808036, PubMed:19111582). Identified in complexes that contain VIM, EZR, AHNAK, BFSP1, BFSP2, ANK2, PLEC, PRX and spectrin (By similarity). Detected in a complex composed of at least EZR, AHNAK, PPL and PRX (By similarity). Interacts with PDPN (via cytoplasmic domain); activates RHOA and promotes epithelial-mesenchymal transition (PubMed:17046996). Interacts with SPN/CD43 cytoplasmic tail, CD44 and ICAM2 (By similarity). Interacts with SLC9A3; interaction targets SLC9A3 to the apical membrane (By similarity). Interacts with SLC9A1; regulates interactions of SLC9A1 with cytoskeletal and promotes stress fiber formation (By similarity). Interacts with CLIC5; may work together in a complex which also includes RDX and MYO6 to stabilize linkages between the plasma membrane and subjacent actin cytoskeleton at the base of stereocilia (By similarity). {ECO:0000250|UniProtKB:P26040, ECO:0000250|UniProtKB:P31976, ECO:0000250|UniProtKB:P31977, ECO:0000250|UniProtKB:Q8HZQ5, ECO:0000269|PubMed:12651155, ECO:0000269|PubMed:12808036, ECO:0000269|PubMed:15498789, ECO:0000269|PubMed:17046996, ECO:0000269|PubMed:17616675, ECO:0000269|PubMed:17881735, ECO:0000269|PubMed:18046454, ECO:0000269|PubMed:19111582, ECO:0000269|PubMed:19555689, ECO:0000269|PubMed:27364155, ECO:0000269|PubMed:9314537}. |
| Post-translational modification PTM: Phosphorylated by tyrosine-protein kinases. Phosphorylation by ROCK2 suppresses the head-to-tail association of the N-terminal and C-terminal halves resulting in an opened conformation which is capable of actin and membrane-binding (By similarity). {ECO:0000250}.; PTM: S-nitrosylation is induced by interferon-gamma and oxidatively-modified low-densitity lipoprotein (LDL(ox)) possibly implicating the iNOS-S100A8/9 transnitrosylase complex. {ECO:0000305|PubMed:25417112}. |
| Domain DOMAIN: Has three main structural domains: an N-terminal FERM domain, a central alpha-helical domain and a C-terminal actin-binding domain. {ECO:0000269|PubMed:27364155}.; DOMAIN: The FERM domain is organized in a clover-shaped structure that comprises three subdomains identified as F1 (residues 2-82), F2 (residues 96-198), and F3 (residues 204-296). In the active form, the subdomain F3 adopts two mutually exclusive conformational isomers where a row of four phenylalanine side chains (Phe250, Phe255, Phe267 and Phe269) must point in the same direction. In the autoinhibited form, the F3 subdomain interacts with the C-terminal domain (residues 516-586) and stabilizes the structure, selecting only one possible arrangement of phenylalanine side chains. The FERM domain mediates binding to membrane lipids and signaling molecules. {ECO:0000269|PubMed:27364155}.; DOMAIN: The central alpha-helical domain is composed of two alpha helices (residues 326-406 and 417-466) connected by a linker. It protrudes from the FERM domain forming a coiled coil structure where the linker can have either a loop or a helix conformation. The monomer is predicted to form an intra-molecular helix-loop-helix coiled coil structure. Whereas the dimer adopts an elongated dumbbell-shaped configuration where continuous alpha helices from each protomer are organized in a antiparallel coiled coil structure that connect FERM:C-terminal domain swapped complex at each end. The dimer is predicted to link actin filaments parallel to the plasma membrane. {ECO:0000305|PubMed:27364155}.; DOMAIN: The [IL]-x-C-x-x-[DE] motif is a proposed target motif for cysteine S-nitrosylation mediated by the iNOS-S100A8/A9 transnitrosylase complex. {ECO:0000305|PubMed:25417112}. |
| Target Relevance information above includes information from UniProt accession: P15311 |
| The UniProt Consortium |
Data
| No results found |
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
| Western blot IHC |
Documents
| # | SDS | Certificate | |
|---|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | |||
Only logged in customers who have purchased this product may leave a review.



Reviews
There are no reviews yet.