| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| host | rabbit |
| isotype | IgG |
| clonality | polyclonal |
| concentration | 1 mg/mL |
| applications | ICC/IF, WB |
| reactivity | NFkB-p65 (Phospho-Ser276) |
| available sizes | 100 µL |
rabbit anti-NFkB-p65 (Phospho-Ser276) polyclonal antibody 8323
$366.00
Antibody summary
- Rabbit polyclonal to NFkB-p65 (Phospho-Ser276)
- Suitable for: WB,IHC
- Isotype: Whole IgG
- 100 µl
rabbit anti-NFkB-p65 (Phospho-Ser276) polyclonal antibody 8323
| antibody |
|---|
| Tested applications WB,IHC,IHC |
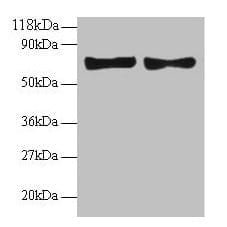
| Recommended dilutions Western blotting: use at dilution of 1:500-1:1,000. A band of ~65kDa is detected. Immunohistochemistry: use at dilution of 1:50-1:100. These are recommended working dilutions. End user should determine optimal dilutions for their applications. |
| Immunogen Peptide sequence that includes phosphorylation site of Serine 276 (R-P-S(p)-D-R) derived from human NFkappaB-p65 and conjugated to KLH. |
| Size and concentration 100µL and 1 mg/mL |
| Form liquid |
| Storage Instructions This antibody is stable for at least one (1) year at -20°C. |
| Storage buffer PBS (without Mg2 and Ca2 ), pH 7.4, 150mM NaCl, |
| Purity affinity purified |
| Clonality polyclonal |
| Isotype IgG |
| Compatible secondaries goat anti-rabbit IgG, H&L chain specific, peroxidase conjugated, conjugated polyclonal antibody 9512 goat anti-rabbit IgG, H&L chain specific, biotin conjugated polyclonal antibody 2079 goat anti-rabbit IgG, H&L chain specific, FITC conjugated polyclonal antibody 7863 goat anti-rabbit IgG, H&L chain specific, Cross Absorbed polyclonal antibody 2371 goat anti-rabbit IgG, H&L chain specific, biotin conjugated polyclonal antibody, crossabsorbed 1715 goat anti-rabbit IgG, H&L chain specific, FITC conjugated polyclonal antibody, crossabsorbed 1720 |
| Isotype control Rabbit polyclonal - Isotype Control |
| target relevance |
|---|
| Protein names Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells 3) |
| Gene names RELA,RELA NFKB3 |
| Mass 60219Da |
| Function FUNCTION: NF-kappa-B is a pleiotropic transcription factor present in almost all cell types and is the endpoint of a series of signal transduction events that are initiated by a vast array of stimuli related to many biological processes such as inflammation, immunity, differentiation, cell growth, tumorigenesis and apoptosis. NF-kappa-B is a homo- or heterodimeric complex formed by the Rel-like domain-containing proteins RELA/p65, RELB, NFKB1/p105, NFKB1/p50, REL and NFKB2/p52. The heterodimeric RELA-NFKB1 complex appears to be most abundant one. The dimers bind at kappa-B sites in the DNA of their target genes and the individual dimers have distinct preferences for different kappa-B sites that they can bind with distinguishable affinity and specificity. Different dimer combinations act as transcriptional activators or repressors, respectively. The NF-kappa-B heterodimeric RELA-NFKB1 and RELA-REL complexes, for instance, function as transcriptional activators. NF-kappa-B is controlled by various mechanisms of post-translational modification and subcellular compartmentalization as well as by interactions with other cofactors or corepressors. NF-kappa-B complexes are held in the cytoplasm in an inactive state complexed with members of the NF-kappa-B inhibitor (I-kappa-B) family. In a conventional activation pathway, I-kappa-B is phosphorylated by I-kappa-B kinases (IKKs) in response to different activators, subsequently degraded thus liberating the active NF-kappa-B complex which translocates to the nucleus. The inhibitory effect of I-kappa-B on NF-kappa-B through retention in the cytoplasm is exerted primarily through the interaction with RELA. RELA shows a weak DNA-binding site which could contribute directly to DNA binding in the NF-kappa-B complex. Beside its activity as a direct transcriptional activator, it is also able to modulate promoters accessibility to transcription factors and thereby indirectly regulate gene expression. Associates with chromatin at the NF-kappa-B promoter region via association with DDX1. Essential for cytokine gene expression in T-cells (PubMed:15790681). The NF-kappa-B homodimeric RELA-RELA complex appears to be involved in invasin-mediated activation of IL-8 expression. Key transcription factor regulating the IFN response during SARS-CoV-2 infection (PubMed:33440148). {ECO:0000269|PubMed:10928981, ECO:0000269|PubMed:12748188, ECO:0000269|PubMed:15790681, ECO:0000269|PubMed:17000776, ECO:0000269|PubMed:17620405, ECO:0000269|PubMed:19058135, ECO:0000269|PubMed:19103749, ECO:0000269|PubMed:20547752, ECO:0000269|PubMed:33440148}. |
| Subellular location SUBCELLULAR LOCATION: Nucleus {ECO:0000269|PubMed:1493333, ECO:0000269|PubMed:15799966, ECO:0000269|PubMed:19058135, ECO:0000269|PubMed:20547752, ECO:0000269|PubMed:25355951}. Cytoplasm {ECO:0000269|PubMed:1493333, ECO:0000269|PubMed:19058135, ECO:0000269|PubMed:20547752, ECO:0000269|PubMed:24807716, ECO:0000269|PubMed:27736973}. Note=Nuclear, but also found in the cytoplasm in an inactive form complexed to an inhibitor (I-kappa-B) (PubMed:1493333). Colocalized with DDX1 in the nucleus upon TNF-alpha induction (PubMed:19058135). Colocalizes with GFI1 in the nucleus after LPS stimulation (PubMed:20547752). Translocation to the nucleus is impaired in L.monocytogenes infection (PubMed:20855622). {ECO:0000269|PubMed:1493333, ECO:0000269|PubMed:19058135, ECO:0000269|PubMed:20547752, ECO:0000269|PubMed:20855622}. |
| Structure SUBUNIT: Component of the NF-kappa-B p65-p50 complex. Component of the NF-kappa-B p65-c-Rel complex. Homodimer; component of the NF-kappa-B p65-p65 complex. Component of the NF-kappa-B p65-p52 complex. May interact with ETHE1. Binds TLE5 and TLE1. Interacts with TP53BP2. Binds to and is phosphorylated by the activated form of either RPS6KA4 or RPS6KA5. Interacts with ING4 and this interaction may be indirect. Interacts with CARM1, USP48 and UNC5CL. Interacts with IRAK1BP1 (By similarity). Interacts with NFKBID (By similarity). Interacts with NFKBIA (PubMed:1493333). Interacts with GSK3B. Interacts with NFKBIB (By similarity). Interacts with NFKBIE. Interacts with NFKBIZ. Interacts with EHMT1 (via ANK repeats) (PubMed:21515635). Part of a 70-90 kDa complex at least consisting of CHUK, IKBKB, NFKBIA, RELA, ELP1 and MAP3K14. Interacts with HDAC3; HDAC3 mediates the deacetylation of RELA. Interacts with HDAC1; the interaction requires non-phosphorylated RELA. Interacts with CBP; the interaction requires phosphorylated RELA. Interacts (phosphorylated at 'Thr-254') with PIN1; the interaction inhibits p65 binding to NFKBIA. Interacts with SOCS1. Interacts with UXT. Interacts with MTDH and PHF11. Interacts with ARRB2. Interacts with NFKBIA (when phosphorylated), the interaction is direct; phosphorylated NFKBIA is part of a SCF(BTRC)-like complex lacking CUL1. Interacts with RNF25. Interacts (via C-terminus) with DDX1. Interacts with UFL1 and COMMD1. Interacts with BRMS1; this promotes deacetylation of 'Lys-310'. Interacts with NOTCH2 (By similarity). Directly interacts with MEN1; this interaction represses NFKB-mediated transactivation. Interacts with AKIP1, which promotes the phosphorylation and nuclear retention of RELA. Interacts (via the RHD) with GFI1; the interaction, after bacterial lipopolysaccharide (LPS) stimulation, inhibits the transcriptional activity by interfering with the DNA-binding activity to target gene promoter DNA. Interacts (when acetylated at Lys-310) with BRD4; leading to activation of the NF-kappa-B pathway. Interacts with MEFV. Interacts with CLOCK (By similarity). Interacts (via N-terminus) with CPEN1; this interaction induces proteolytic cleavage of p65/RELA subunit and inhibition of NF-kappa-B transcriptional activity (PubMed:18212740). Interacts with FOXP3. Interacts with CDK5RAP3; stimulates the interaction of RELA with HDAC1, HDAC2 and HDAC3 thereby inhibiting NF-kappa-B transcriptional activity (PubMed:17785205). Interacts with DHX9; this interaction is direct and activates NF-kappa-B-mediated transcription (PubMed:15355351). Interacts with LRRC25 (PubMed:29044191). Interacts with TBX21 (By similarity). Interacts with KAT2A (By similarity). Interacts with ZBTB7A; involved in the control by RELA of the accessibility of target gene promoters (PubMed:29813070). Directly interacts with DDX3X; this interaction may trap RELA in the cytoplasm, impairing nuclear relocalization upon TNF activating signals (PubMed:27736973). Interacts with PHF2 (By similarity). Interacts with MKRN2; the interaction leads to its polyubiquitination and proteasome-dependent degradation (By similarity). Interacts with ECSIT (PubMed:25355951). {ECO:0000250|UniProtKB:Q04207, ECO:0000269|PubMed:1493333, ECO:0000269|PubMed:15355351, ECO:0000269|PubMed:15790681, ECO:0000269|PubMed:17785205, ECO:0000269|PubMed:18212740, ECO:0000269|PubMed:21515635, ECO:0000269|PubMed:25355951, ECO:0000269|PubMed:27736973, ECO:0000269|PubMed:29044191, ECO:0000269|PubMed:29813070}.; SUBUNIT: (Microbial infection) Interacts with human respiratory syncytial virus (HRSV) protein M2-1. {ECO:0000269|PubMed:15629770}.; SUBUNIT: (Microbial infection) Interacts with molluscum contagiosum virus MC132. {ECO:0000269|PubMed:26041281}.; SUBUNIT: (Microbial infection) Interacts with herpes virus 8 virus protein LANA1. {ECO:0000269|PubMed:21697472}.; SUBUNIT: (Microbial infection) Interacts with human cytomegalovirus protein UL44; this interaction prevents NF-kappa-B binding to its promoters. {ECO:0000269|PubMed:30867312}. |
| Post-translational modification PTM: Ubiquitinated by RNF182, leading to its proteasomal degradation (PubMed:31432514). Degradation is required for termination of NF-kappa-B response. Polyubiquitinated via 'Lys-29'-linked ubiquitin; leading to lysosomal degradation (PubMed:21518757). {ECO:0000269|PubMed:15226358, ECO:0000269|PubMed:21518757, ECO:0000269|PubMed:31432514}.; PTM: Monomethylated at Lys-310 by SETD6 (PubMed:21515635). Monomethylation at Lys-310 is recognized by the ANK repeats of EHMT1 and promotes the formation of repressed chromatin at target genes, leading to down-regulation of NF-kappa-B transcription factor activity. Phosphorylation at Ser-311 disrupts the interaction with EHMT1 without preventing monomethylation at Lys-310 and relieves the repression of target genes (By similarity). {ECO:0000250|UniProtKB:Q04207, ECO:0000269|PubMed:21515635}.; PTM: Phosphorylation at Ser-311 disrupts the interaction with EHMT1 and promotes transcription factor activity (By similarity). Phosphorylation on Ser-536 stimulates acetylation on Lys-310 and interaction with CBP; the phosphorylated and acetylated forms show enhanced transcriptional activity. Phosphorylation at Ser-276 by RPS6KA4 and RPS6KA5 promotes its transactivation and transcriptional activities. {ECO:0000250, ECO:0000269|PubMed:10521409, ECO:0000269|PubMed:10938077, ECO:0000269|PubMed:11931769, ECO:0000269|PubMed:12456660, ECO:0000269|PubMed:12628924, ECO:0000269|PubMed:14690596, ECO:0000269|PubMed:15073167, ECO:0000269|PubMed:15516339, ECO:0000269|PubMed:15775976, ECO:0000269|PubMed:16046471, ECO:0000269|PubMed:16135789, ECO:0000269|PubMed:16407239, ECO:0000269|PubMed:17000776, ECO:0000269|PubMed:19103749}.; PTM: Phosphorylation at Ser-75 by herpes simplex virus 1/HHV-1 inhibits NF-kappa-B activation. {ECO:0000269|PubMed:24807716}.; PTM: Reversibly acetylated; the acetylation seems to be mediated by CBP, the deacetylation by HDAC3 and SIRT2. Acetylation at Lys-122 enhances DNA binding and impairs association with NFKBIA. Acetylation at Lys-310 is required for full transcriptional activity in the absence of effects on DNA binding and NFKBIA association. Acetylation at Lys-310 promotes interaction with BRD4. Acetylation can also lower DNA-binding and results in nuclear export. Interaction with BRMS1 promotes deacetylation of Lys-310. Lys-310 is deacetylated by SIRT2. {ECO:0000269|PubMed:12419806, ECO:0000269|PubMed:12456660, ECO:0000269|PubMed:16135789, ECO:0000269|PubMed:17000776, ECO:0000269|PubMed:19103749}.; PTM: S-nitrosylation of Cys-38 inactivates the enzyme activity. {ECO:0000250}.; PTM: Sulfhydration at Cys-38 mediates the anti-apoptotic activity by promoting the interaction with RPS3 and activating the transcription factor activity. {ECO:0000250}.; PTM: Sumoylation by PIAS3 negatively regulates DNA-bound activated NF-kappa-B. {ECO:0000269|PubMed:22649547}.; PTM: Proteolytically cleaved within a conserved N-terminus region required for base-specific contact with DNA in a CPEN1-mediated manner, and hence inhibits NF-kappa-B transcriptional activity (PubMed:18212740). {ECO:0000269|PubMed:18212740}. |
| Domain DOMAIN: The transcriptional activation domain 3/TA3 does not participate in the direct transcriptional activity of RELA but is involved in the control by RELA of the accessibility of target gene promoters. Mediates interaction with ZBTB7A. {ECO:0000250|UniProtKB:Q04207}.; DOMAIN: The transcriptional activation domain 1/TA1 and the transcriptional activation domain 2/TA2 have direct transcriptional activation properties (By similarity). The 9aaTAD motif found within the transcriptional activation domain 2 is a conserved motif present in a large number of transcription factors that is required for their transcriptional transactivation activity (PubMed:17467953). {ECO:0000250|UniProtKB:Q04207, ECO:0000269|PubMed:17467953}. |
| Involvement in disease DISEASE: Note=A chromosomal aberration involving ZFTA is found in more than two-thirds of supratentorial ependymomas. Translocation with ZFTA produces a ZFTA-RELA fusion protein. ZFTA-RELA translocations are potent oncogenes that probably transform neural stem cells by driving an aberrant NF-kappa-B transcription program (PubMed:24553141). {ECO:0000269|PubMed:24553141}.; DISEASE: Autoinflammatory disease, familial, Behcet-like 3 (AIFBL3) [MIM:618287]: An autosomal dominant, mucocutaneous disease characterized by chronic mucosal lesions, in absence of recurrent infections. {ECO:0000269|PubMed:28600438}. Note=The disease may be caused by variants affecting the gene represented in this entry. |
| Target Relevance information above includes information from UniProt accession: Q04206 |
| The UniProt Consortium |
Data
| No results found |
Publications
Warning: Cannot modify header information - headers already sent by (output started at /www/benchmarkantibodiescom_769/public/wp-includes/script-loader.php:3015) in /www/benchmarkantibodiescom_769/public/wp-content/plugins/shortcode-manager/shortcode-manager.php(453) : eval()'d code on line 3
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
| Western blot IHC |
Documents
| # | SDS | Certificate | |
|---|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | |||
Only logged in customers who have purchased this product may leave a review.



Reviews
There are no reviews yet.