| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| host | rabbit |
| isotype | IgG |
| clonality | polyclonal |
| concentration | 1 mg/mL |
| applications | ICC/IF, WB |
| reactivity | MAP-1 |
| available sizes | 100 µg |
rabbit anti-MAP-1 polyclonal antibody 6884
$376.00
Antibody summary
- Rabbit polyclonal to MAP-1
- Suitable for: ELISA
- Isotype: Whole IgG
- 100 µg
rabbit anti-MAP-1 polyclonal antibody 6884
| antibody |
|---|
| Tested applications ELISA |
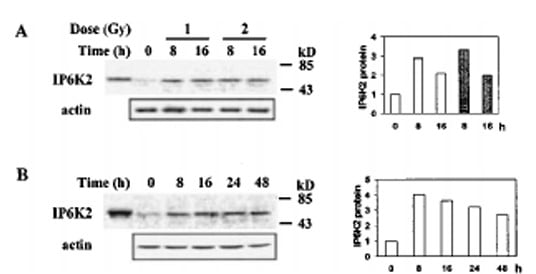
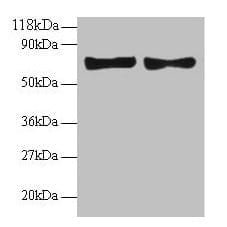
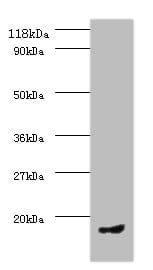
| Recommended dilutions Immunoblotting: use at 1:500-1:1,000 dilution. A band of approx. 39kDa is detected. Positive control: Heart or brain tissue lysate. These are recommended concentrations. Enduser should determine optimal concentrations for their applications. |
| Immunogen Synthetic peptide corresponding to aa 337-351 of human MAP-1. |
| Size and concentration 100µg and lot specific |
| Form liquid |
| Storage Instructions This antibody is stable for at least one (1) year at -20°C. Avoid multiple freeze-thaw cycles. |
| Storage buffer PBS, pH 7.4. |
| Purity peptide affinity purification |
| Clonality polyclonal |
| Isotype IgG |
| Compatible secondaries goat anti-rabbit IgG, H&L chain specific, peroxidase conjugated, conjugated polyclonal antibody 9512 goat anti-rabbit IgG, H&L chain specific, biotin conjugated polyclonal antibody 2079 goat anti-rabbit IgG, H&L chain specific, FITC conjugated polyclonal antibody 7863 goat anti-rabbit IgG, H&L chain specific, Cross Absorbed polyclonal antibody 2371 goat anti-rabbit IgG, H&L chain specific, biotin conjugated polyclonal antibody, crossabsorbed 1715 goat anti-rabbit IgG, H&L chain specific, FITC conjugated polyclonal antibody, crossabsorbed 1720 |
| Isotype control Rabbit polyclonal - Isotype Control |
| target relevance |
|---|
| Protein names Modulator of apoptosis 1 (MAP-1) (MAP1) (Paraneoplastic antigen Ma4) |
| Gene names MOAP1,MOAP1 PNMA4 |
| Protein family PNMA family |
| Mass 39513Da |
| Function FUNCTION: Retrotransposon-derived protein that forms virion-like capsids (By similarity). Acts as an effector of BAX during apoptosis: enriched at outer mitochondria membrane and associates with BAX upon induction of apoptosis, facilitating BAX-dependent mitochondrial outer membrane permeabilization and apoptosis (PubMed:11060313, PubMed:16199525). Required for death receptor-dependent apoptosis (PubMed:11060313). When associated with RASSF1, promotes BAX conformational change and translocation to mitochondrial membranes in response to TNF and TNFSF10 stimulation (PubMed:15949439). Also promotes autophagy: promotes phagophore closure via association with ATG8 proteins (PubMed:33783314). Acts as an inhibitor of the NFE2L2/NRF2 pathway via interaction with SQSTM1: interaction promotes dissociation of SQSTM1 inclusion bodies that sequester KEAP1, relieving inactivation of the BCR(KEAP1) complex (PubMed:33393215). {ECO:0000250|UniProtKB:Q9ERH6, ECO:0000269|PubMed:11060313, ECO:0000269|PubMed:15949439, ECO:0000269|PubMed:16199525, ECO:0000269|PubMed:33393215, ECO:0000269|PubMed:33783314}. |
| Subellular location SUBCELLULAR LOCATION: Cytoplasm, cytosol {ECO:0000269|PubMed:19100260}. Mitochondrion outer membrane {ECO:0000269|PubMed:16199525}. Extracellular vesicle membrane {ECO:0000250|UniProtKB:Q9ERH6}. Note=Forms virion-like extracellular vesicles that are released from cells. {ECO:0000250|UniProtKB:Q9ERH6}. |
| Tissues TISSUE SPECIFICITY: Widely expressed, with high levels in heart and brain. {ECO:0000269|PubMed:19366867}. |
| Structure SUBUNIT: Homodimer (PubMed:15949439). Under normal circumstances, held in an inactive conformation by an intramolecular interaction (PubMed:15949439). Interacts with BAX (PubMed:11060313, PubMed:16199525). Binding to RASSF1 isoform A (RASSF1A) relieves this inhibitory interaction and allows further binding to BAX (PubMed:15949439). Also binds to BCL2 and BCLX (PubMed:11060313). Recruited to the TNFRSF1A and TNFRSF10A complexes in response to their respective cognate ligand, after internalization (PubMed:15949439). Interacts with TRIM39 (PubMed:19100260). Interacts with RASSF6 (By similarity). Interacts with ATG8 proteins MAP1LC3A, MAP1LC3B and MAP1LC3C (PubMed:33783314). Does not interact with ATG8 proteins GABARAPL1, GABARAPL2 and GABARAP (PubMed:33783314). Interacts with SQSTM1; promoting dissociation of SQSTM1 inclusion bodies that sequester KEAP1 (PubMed:33393215). {ECO:0000250|UniProtKB:Q9ERH6, ECO:0000269|PubMed:11060313, ECO:0000269|PubMed:15949439, ECO:0000269|PubMed:16199525, ECO:0000269|PubMed:19100260, ECO:0000269|PubMed:33393215, ECO:0000269|PubMed:33783314}. |
| Post-translational modification PTM: Ubiquitinated and degraded during mitotic exit by APC/C-Cdh1, this modification is inhibited by TRIM39. {ECO:0000269|PubMed:22529100}. |
| Domain DOMAIN: The protein is evolutionarily related to retrotransposon Gag proteins: it contains the capsid (CA)subdomain of gag. {ECO:0000269|PubMed:34357660}.; DOMAIN: The BH3-like domain is required for association with BAX and for mediating apoptosis (PubMed:11060313). The three BH domains (BH1, BH2, and BH3) of BAX are all required for mediating protein-protein interaction (PubMed:11060313). {ECO:0000269|PubMed:11060313}.; DOMAIN: The LIR motif (LC3-interacting region) is required for the interaction with the ATG8 family proteins MAP1LC3A, MAP1LC3B and MAP1LC3C. {ECO:0000269|PubMed:33783314}. |
| Target Relevance information above includes information from UniProt accession: Q96BY2 |
| The UniProt Consortium |
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
| ELISA |
Documents
| # | SDS | Certificate | |
|---|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | |||
Only logged in customers who have purchased this product may leave a review.



Reviews
There are no reviews yet.