| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| host | rabbit |
| isotype | IgG |
| clonality | polyclonal |
| concentration | 1 mg/mL |
| applications | ICC/IF, WB |
| reactivity | CDC25a |
| available sizes | 100 µg |
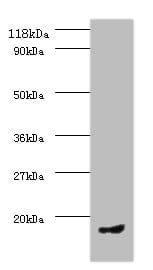
rabbit anti-CDC25a polyclonal antibody 8227
$518.00
Antibody summary
- Rabbit polyclonal to CDC25a
- Suitable for: WB,IP
- Isotype: Whole IgG
- 100 µg
rabbit anti-CDC25a polyclonal antibody 8227
| antibody |
|---|
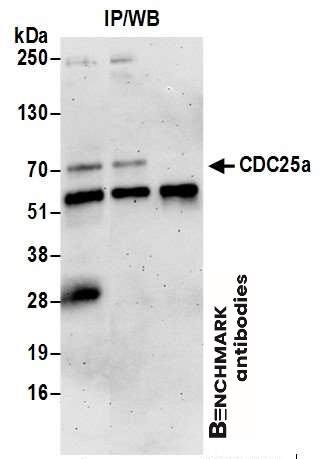
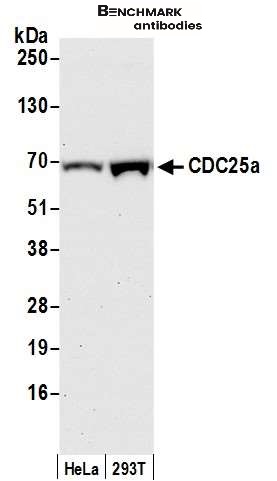
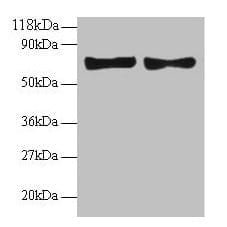
| Tested applications WB,IP |
| Recommended dilutions Western Blot: 0.1 - 1 ug/ml. Immunoprecipitation: 1- 4 ug/mg of lysate. |
| Immunogen Synthetic peptide representing a portion of the protein encoded by exon 7 (LocusLink ID 993). |
| Size and concentration 100µg and lot specific |
| Form liquid |
| Storage Instructions Store at 2 - 8°C. Antibody is stable at 2 - 8°C for 1 year. |
| Storage buffer Tris-citrate/phosphate buffer, pH 7 to 8 contai |
| Purity immunogen affinity purification |
| Clonality polyclonal |
| Isotype IgG |
| Compatible secondaries goat anti-rabbit IgG, H&L chain specific, peroxidase conjugated, conjugated polyclonal antibody 9512 goat anti-rabbit IgG, H&L chain specific, biotin conjugated polyclonal antibody 2079 goat anti-rabbit IgG, H&L chain specific, FITC conjugated polyclonal antibody 7863 goat anti-rabbit IgG, H&L chain specific, Cross Absorbed polyclonal antibody 2371 goat anti-rabbit IgG, H&L chain specific, biotin conjugated polyclonal antibody, crossabsorbed 1715 goat anti-rabbit IgG, H&L chain specific, FITC conjugated polyclonal antibody, crossabsorbed 1720 |
| Isotype control Rabbit polyclonal - Isotype Control |
| target relevance |
|---|
| Protein names M-phase inducer phosphatase 1 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25A) |
| Gene names CDC25A,CDC25A |
| Protein family MPI phosphatase family |
| Mass 59087Da |
| Function FUNCTION: Tyrosine protein phosphatase which functions as a dosage-dependent inducer of mitotic progression (PubMed:12676925, PubMed:14559997, PubMed:1836978, PubMed:20360007). Directly dephosphorylates CDK1 and stimulates its kinase activity (PubMed:20360007). Also dephosphorylates CDK2 in complex with cyclin-E, in vitro (PubMed:20360007). {ECO:0000269|PubMed:12676925, ECO:0000269|PubMed:14559997, ECO:0000269|PubMed:1836978, ECO:0000269|PubMed:20360007}. |
| Catalytic activity CATALYTIC ACTIVITY: Reaction=O-phospho-L-tyrosyl-[protein] + H2O = L-tyrosyl-[protein] + phosphate; Xref=Rhea:RHEA:10684, Rhea:RHEA-COMP:10136, Rhea:RHEA-COMP:20101, ChEBI:CHEBI:15377, ChEBI:CHEBI:43474, ChEBI:CHEBI:46858, ChEBI:CHEBI:61978; EC=3.1.3.48; Evidence={ECO:0000269|PubMed:12676925, ECO:0000269|PubMed:14559997, ECO:0000305|PubMed:20360007}; PhysiologicalDirection=left-to-right; Xref=Rhea:RHEA:10685; Evidence={ECO:0000305|PubMed:20360007}; |
| Structure SUBUNIT: Interacts with CCNB1/cyclin B1. Interacts with YWHAE/14-3-3 epsilon when phosphorylated. Interacts with CUL1 specifically when CUL1 is neddylated and active. Interacts with BTRC/BTRCP1 and FBXW11/BTRCP2. Interactions with CUL1, BTRC and FBXW11 are enhanced upon DNA damage. Interacts with CHEK2; mediates CDC25A phosphorylation and degradation in response to infrared-induced DNA damages. Interacts with HSP90AB1; prevents heat shock-mediated CDC25A degradation and contributes to cell cycle progression (PubMed:22843495). {ECO:0000269|PubMed:11298456, ECO:0000269|PubMed:14559997, ECO:0000269|PubMed:14681206, ECO:0000269|PubMed:16356754, ECO:0000269|PubMed:22843495}. |
| Post-translational modification PTM: Phosphorylated by CHEK1 on Ser-76, Ser-124, Ser-178, Ser-279, Ser-293 and Thr-507 during checkpoint mediated cell cycle arrest. Also phosphorylated by CHEK2 on Ser-124, Ser-279, and Ser-293 during checkpoint mediated cell cycle arrest. Phosphorylation on Ser-178 and Thr-507 creates binding sites for YWHAE/14-3-3 epsilon which inhibits CDC25A. Phosphorylation on Ser-76, Ser-124, Ser-178, Ser-279 and Ser-293 may also promote ubiquitin-dependent proteolysis of CDC25A by the SCF complex. Phosphorylation of CDC25A at Ser-76 by CHEK1 primes it for subsequent phosphorylation at Ser-79, Ser-82 and Ser-88 by NEK11. Phosphorylation by NEK11 is required for BTRC-mediated polyubiquitination and degradation. Phosphorylation by PIM1 leads to an increase in phosphatase activity. Phosphorylated by PLK3 following DNA damage, leading to promote its ubiquitination and degradation. {ECO:0000269|PubMed:11298456, ECO:0000269|PubMed:12399544, ECO:0000269|PubMed:12676583, ECO:0000269|PubMed:12676925, ECO:0000269|PubMed:12759351, ECO:0000269|PubMed:14559997, ECO:0000269|PubMed:14681206, ECO:0000269|PubMed:19734889, ECO:0000269|PubMed:20090422, ECO:0000269|PubMed:20228808, ECO:0000269|PubMed:21376736}.; PTM: Ubiquitinated by the anaphase promoting complex/cyclosome (APC/C) ubiquitin ligase complex that contains FZR1/CDH1 during G1 phase leading to its degradation by the proteasome. Ubiquitinated by a SCF complex containing BTRC and FBXW11 during S phase leading to its degradation by the proteasome. Deubiquitination by USP17L2/DUB3 leads to its stabilization. {ECO:0000269|PubMed:20228808}. |
| Domain DOMAIN: The phosphodegron motif mediates interaction with specific F-box proteins when phosphorylated. Putative phosphorylation sites at Ser-79 and Ser-82 appear to be essential for this interaction. {ECO:0000269|PubMed:12234927}. |
| Target Relevance information above includes information from UniProt accession: P30304 |
| The UniProt Consortium |
Data
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
| Western blot |
Documents
| # | SDS | Certificate | |
|---|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | |||
Only logged in customers who have purchased this product may leave a review.





Reviews
There are no reviews yet.