| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| host | mouse |
| isotype | IgG2a |
| clonality | monoclonal |
| concentration | concentrate, predilute |
| applications | IHC |
| reactivity | human |
| available size | 0.1 mL, 0.5 mL, 1 mL concentrated, 7 mL prediluted |
mouse anti-PCNA monoclonal antibody (PC10) 6325
Price range: $160.00 through $528.00
Antibody summary
- Mouse monoclonal to PCNA
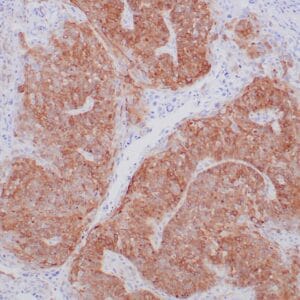
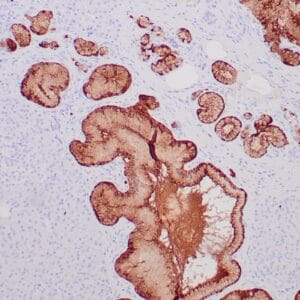
- Suitable for: Immunohistochemistry (formalin-fixed, paraffin-embedded tissues)
- Reacts with: Human
- Isotype:IgG2a
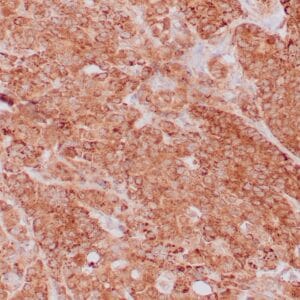
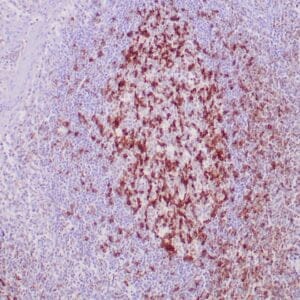
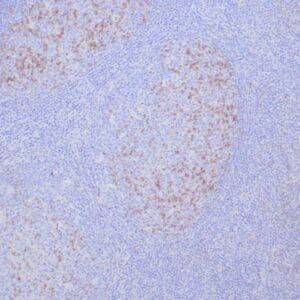
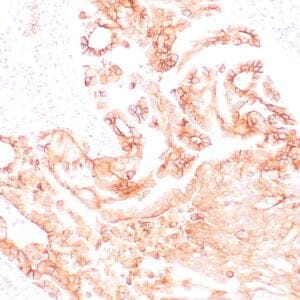
- Control: Tonsil, lymph node or small intestine
- Visualization: Nuclear
- 0.1, 0.5, 1.0 mL concentrated, 7 mL prediluted
mouse anti-PCNA monoclonal antibody PC10 6325
| target relevance |
|---|
| Protein names Proliferating cell nuclear antigen (PCNA) (Cyclin) |
| Gene names PCNA,PCNA |
| Protein family PCNA family |
| Mass 28769Da |
| Function FUNCTION: Auxiliary protein of DNA polymerase delta and epsilon, is involved in the control of eukaryotic DNA replication by increasing the polymerase's processibility during elongation of the leading strand (PubMed:35585232). Induces a robust stimulatory effect on the 3'-5' exonuclease and 3'-phosphodiesterase, but not apurinic-apyrimidinic (AP) endonuclease, APEX2 activities. Has to be loaded onto DNA in order to be able to stimulate APEX2. Plays a key role in DNA damage response (DDR) by being conveniently positioned at the replication fork to coordinate DNA replication with DNA repair and DNA damage tolerance pathways (PubMed:24939902). Acts as a loading platform to recruit DDR proteins that allow completion of DNA replication after DNA damage and promote postreplication repair: Monoubiquitinated PCNA leads to recruitment of translesion (TLS) polymerases, while 'Lys-63'-linked polyubiquitination of PCNA is involved in error-free pathway and employs recombination mechanisms to synthesize across the lesion (PubMed:24695737). {ECO:0000269|PubMed:18719106, ECO:0000269|PubMed:19443450, ECO:0000269|PubMed:24695737, ECO:0000269|PubMed:24939902, ECO:0000269|PubMed:35585232, ECO:0000269|PubMed:38459011}. |
| Subellular location SUBCELLULAR LOCATION: Nucleus {ECO:0000269|PubMed:15543136, ECO:0000269|PubMed:24115439, ECO:0000269|PubMed:24939902, ECO:0000269|PubMed:38459011}. Note=Colocalizes with CREBBP, EP300 and POLD1 to sites of DNA damage (PubMed:24939902). Forms nuclear foci representing sites of ongoing DNA replication and vary in morphology and number during S phase (PubMed:15543136). Co-localizes with SMARCA5/SNF2H and BAZ1B/WSTF at replication foci during S phase (PubMed:15543136). Together with APEX2, is redistributed in discrete nuclear foci in presence of oxidative DNA damaging agents. {ECO:0000269|PubMed:15543136, ECO:0000269|PubMed:24939902}. |
| Structure SUBUNIT: Homotrimer (PubMed:24939902). Interacts with p300/EP300; the interaction occurs on chromatin in UV-irradiated damaged cells (PubMed:24939902). Interacts with CREBBP (via transactivation domain and C-terminus); the interaction occurs on chromatin in UV-irradiated damaged cells (PubMed:24939902). Directly interacts with POLD1, POLD3 and POLD4 subunits of the DNA polymerase delta complex, POLD3 being the major interacting partner; the interaction with POLD3 is inhibited by CDKN1A/p21(CIP1) (PubMed:11595739, PubMed:16510448, PubMed:22148433, PubMed:24939902). Forms a complex with activator 1 heteropentamer in the presence of ATP. Interacts with EXO1, POLH, POLK, DNMT1, ERCC5, FEN1, CDC6 and POLDIP2 (PubMed:11784855, PubMed:12522211, PubMed:15149598, PubMed:15225546, PubMed:15616578, PubMed:24911150, PubMed:26760506, PubMed:9302295, PubMed:9305916, PubMed:9566895). Interacts with POLB (PubMed:19336415, PubMed:26760506). Interacts with APEX2; this interaction is triggered by reactive oxygen species and increased by misincorporation of uracil in nuclear DNA (PubMed:11376153, PubMed:19443450). Forms a ternary complex with DNTTIP2 and core histone (PubMed:12786946). Interacts with KCTD10 and PPP1R15A (By similarity). Interacts with SMARCA5/SNF2H (PubMed:15543136). Interacts with BAZ1B/WSTF; the interaction is direct and is required for BAZ1B/WSTF binding to replication foci during S phase (PubMed:15543136). Interacts with HLTF and SHPRH (PubMed:17130289, PubMed:18316726, PubMed:18719106). Interacts with NUDT15; this interaction is disrupted in response to UV irradiation and acetylation (PubMed:19419956). Interacts with CDKN1A/p21(CIP1) and CDT1; interacts via their PIP-box which also recruits the DCX(DTL) complex. The interaction with CDKN1A inhibits POLD3 binding (PubMed:11595739, PubMed:16949367, PubMed:18703516, PubMed:18794347). Interacts with DDX11 (PubMed:18499658). Interacts with EGFR; positively regulates PCNA (PubMed:17115032). Interacts with PARPBP (PubMed:22153967). Interacts (when ubiquitinated) with SPRTN; leading to enhance RAD18-mediated PCNA ubiquitination (PubMed:22681887, PubMed:27084448). Interacts (when polyubiquitinated) with ZRANB3 (PubMed:22704558, PubMed:22705370, PubMed:22759634). Interacts with SMARCAD1 (PubMed:21549307). Interacts with CDKN1C (PubMed:22634751). Interacts with PCLAF (via PIP-box) (PubMed:21628590, PubMed:23000965). Interacts with RTEL1 (via PIP-box); the interaction is direct and essential for the suppression of telomere fragility (PubMed:24115439). Interacts with FAM111A (via PIP-box); the interaction is direct and required for PCNA loading on chromatin binding (PubMed:24561620). Interacts with LIG1 (PubMed:24911150). Interacts with SETMAR (PubMed:20457750). Interacts with ANKRD17 (PubMed:23711367). Interacts with FBXO18/FBH1 (via PIP-box); the interaction recruits the DCX(DTL) complex and promotes ubiquitination and degradation of FBXO18/FBH1 (PubMed:23677613). Interacts with POLN (PubMed:19995904). Interacts with SDE2 (via PIP-box); the interaction is direct and prevents ultraviolet light induced monoubiquitination (PubMed:27906959). Component of the replisome complex composed of at least DONSON, MCM2, MCM7, PCNA and TICRR; interaction at least with PCNA occurs during DNA replication (PubMed:28191891). Interacts with MAPK15; the interaction is chromatin binding dependent and prevents MDM2-mediated PCNA destruction by inhibiting the association of PCNA with MDM2 (PubMed:20733054). Interacts with PARP10 (via PIP-box) (PubMed:24695737). Interacts with DDI2 (PubMed:29290612). Interacts with HMCES (via PIP-box) (PubMed:30554877). Interacts with TRAIP (via PIP-box) (PubMed:26711499, PubMed:27462463). Interacts with UHRF2 (PubMed:28951215). Interacts with ALKBH2; this interaction is enhanced during the S-phase of the cell cycle. Interacts with ATAD5; the interaction promotes USP1-mediated PCNA deubiquitination (PubMed:20147293). Interacts with DNA damage up-regulated protein DDUP (PubMed:35849344). Interacts (when phosphorylated) with GRB2 (PubMed:38459011). Interacts with ANG (PubMed:37218877). Interacts with nuclear UNG (isoform 2); this interaction mediates UNG recruitment to S-phase replication foci. {ECO:0000250|UniProtKB:P04961, ECO:0000250|UniProtKB:P17918, ECO:0000269|PubMed:11376153, ECO:0000269|PubMed:11595739, ECO:0000269|PubMed:11784855, ECO:0000269|PubMed:12522211, ECO:0000269|PubMed:12786946, ECO:0000269|PubMed:15149598, ECO:0000269|PubMed:15225546, ECO:0000269|PubMed:15543136, ECO:0000269|PubMed:15616578, ECO:0000269|PubMed:16510448, ECO:0000269|PubMed:16949367, ECO:0000269|PubMed:17115032, ECO:0000269|PubMed:17130289, ECO:0000269|PubMed:18316726, ECO:0000269|PubMed:18499658, ECO:0000269|PubMed:18703516, ECO:0000269|PubMed:18719106, ECO:0000269|PubMed:18794347, ECO:0000269|PubMed:19336415, ECO:0000269|PubMed:19419956, ECO:0000269|PubMed:19443450, ECO:0000269|PubMed:19995904, ECO:0000269|PubMed:20147293, ECO:0000269|PubMed:20457750, ECO:0000269|PubMed:20733054, ECO:0000269|PubMed:21549307, ECO:0000269|PubMed:21628590, ECO:0000269|PubMed:22148433, ECO:0000269|PubMed:22153967, ECO:0000269|PubMed:22521144, ECO:0000269|PubMed:22634751, ECO:0000269|PubMed:22681887, ECO:0000269|PubMed:22704558, ECO:0000269|PubMed:22705370, ECO:0000269|PubMed:22759634, ECO:0000269|PubMed:23000965, ECO:0000269|PubMed:23677613, ECO:0000269|PubMed:23711367, ECO:0000269|PubMed:24115439, ECO:0000269|PubMed:24561620, ECO:0000269|PubMed:24695737, ECO:0000269|PubMed:24911150, ECO:0000269|PubMed:24939902, ECO:0000269|PubMed:26408825, ECO:0000269|PubMed:26760506, ECO:0000269|PubMed:27084448, ECO:0000269|PubMed:27462463, ECO:0000269|PubMed:27906959, ECO:0000269|PubMed:28191891, ECO:0000269|PubMed:29290612, ECO:0000269|PubMed:30554877, ECO:0000269|PubMed:35849344, ECO:0000269|PubMed:37218877, ECO:0000269|PubMed:38459011, ECO:0000269|PubMed:9302295, ECO:0000269|PubMed:9305916, ECO:0000269|PubMed:9566895}.; SUBUNIT: (Microbial infection) Interacts with herpes virus 8 protein LANA1. {ECO:0000269|PubMed:26223641}. |
| Post-translational modification PTM: Phosphorylated. Phosphorylation at Tyr-211 by EGFR stabilizes chromatin-associated PCNA. {ECO:0000269|PubMed:17115032}.; PTM: Acetylated by CREBBP and p300/EP300; preferentially acetylated by CREBBP on Lys-80, Lys-13 and Lys-14 and on Lys-77 by p300/EP300 upon loading on chromatin in response to UV irradiation (PubMed:19419956, PubMed:24939902). Lysine acetylation disrupts association with chromatin, hence promoting PCNA ubiquitination and proteasomal degradation in response to UV damage in a CREBBP- and EP300-dependent manner (PubMed:24939902). Acetylation disrupts interaction with NUDT15 and promotes degradation (PubMed:19419956). {ECO:0000269|PubMed:24939902}.; PTM: Ubiquitinated (PubMed:20227374, PubMed:24939902). Following DNA damage, can be either monoubiquitinated to stimulate direct bypass of DNA lesions by specialized DNA polymerases or polyubiquitinated to promote recombination-dependent DNA synthesis across DNA lesions by template switching mechanisms. Following induction of replication stress, monoubiquitinated by the UBE2B-RAD18 complex on Lys-164, leading to recruit translesion (TLS) polymerases, which are able to synthesize across DNA lesions in a potentially error-prone manner. An error-free pathway also exists and requires non-canonical polyubiquitination on Lys-164 through 'Lys-63' linkage of ubiquitin moieties by the E2 complex UBE2N-UBE2V2 and the E3 ligases, HLTF, RNF8 and SHPRH. This error-free pathway, also known as template switching, employs recombination mechanisms to synthesize across the lesion, using as a template the undamaged, newly synthesized strand of the sister chromatid. Monoubiquitination at Lys-164 also takes place in undamaged proliferating cells, and is mediated by the DCX(DTL) complex, leading to enhance PCNA-dependent translesion DNA synthesis. Sumoylated during S phase. {ECO:0000269|PubMed:15149598, ECO:0000269|PubMed:17108083, ECO:0000269|PubMed:17130289, ECO:0000269|PubMed:18316726, ECO:0000269|PubMed:18719106, ECO:0000269|PubMed:18948756, ECO:0000269|PubMed:20129063, ECO:0000269|PubMed:20227374, ECO:0000269|PubMed:22153967, ECO:0000269|PubMed:24939902}.; PTM: Methylated on glutamate residues by ARMT1/C6orf211. {ECO:0000269|PubMed:25732820}. |
| Involvement in disease DISEASE: Ataxia-telangiectasia-like disorder 2 (ATLD2) [MIM:615919]: A neurodegenerative disorder due to defects in DNA excision repair. ATLD2 is characterized by developmental delay, ataxia, sensorineural hearing loss, short stature, cutaneous and ocular telangiectasia, and photosensitivity. {ECO:0000269|PubMed:24911150}. Note=The disease is caused by variants affecting the gene represented in this entry. |
| Target Relevance information above includes information from UniProt accession: P12004 |
| The UniProt Consortium |
Data
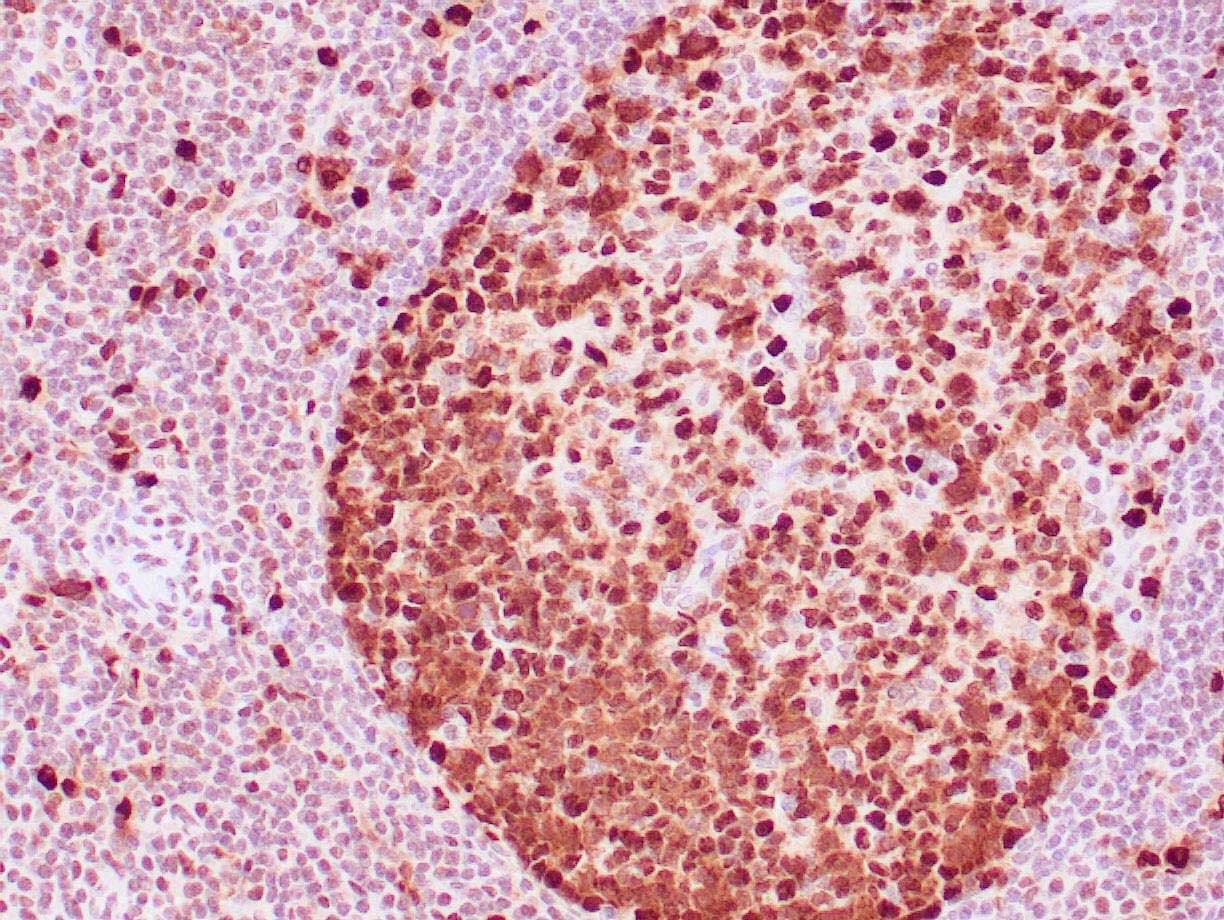 |
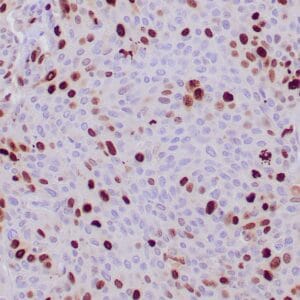
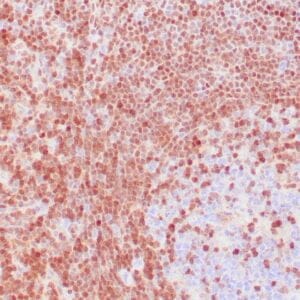
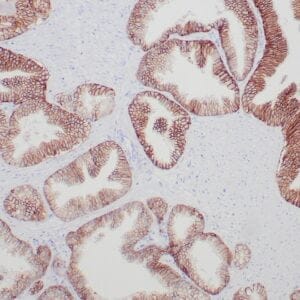
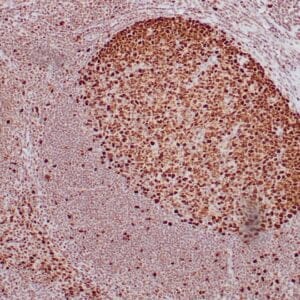
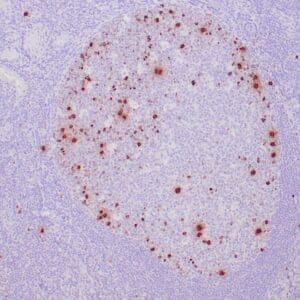
| Human tonsil stained with anti-PCNA antibody using peroxidase-conjugate and DAB chromogen. Note the nuclear staining of lymphoid cells. |
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
| IHC |
Documents
| # | SDS | Certificate | |
|---|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | |||
Only logged in customers who have purchased this product may leave a review.

Reviews
There are no reviews yet.