| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| host | mouse |
| isotype | IgG1 |
| clonality | monoclonal |
| concentration | 1 mg/mL |
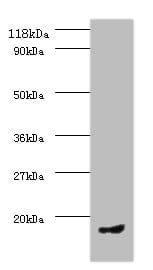
| applications | ICC/IF, WB |
| reactivity | p53 |
| available sizes | 100 µg |
mouse anti-p53 monoclonal antibody (240) 5777
$503.00
Antibody summary
- Mouse monoclonal to p53
- Suitable for: WB,ICC/IF,IHC-P,IHC-Fr,FACS,IP,ELISA
- Isotype: IgG1
- 100 µg
mouse anti-p53 monoclonal antibody (240) 5777
| antibody |
|---|
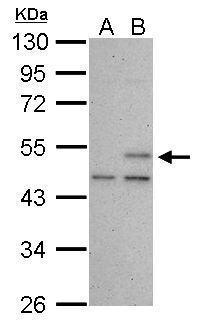
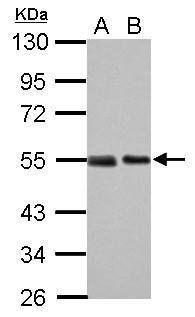
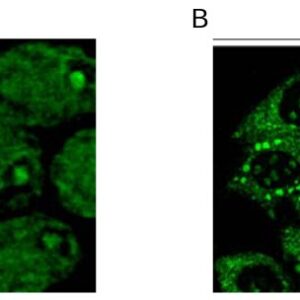
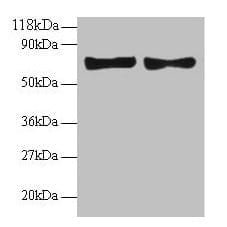
| Tested applications WB,IHC,IHC,ICC/IF |
| Recommended dilutions ELISA, FACS, IHC (frozen and paraffin), Immunoprecipitation, Immunoblotting. IHC: use at 2-4ug/ml. Extensive washing may be necessary. Protein digestion and/or microwave antigen retrieval prior to staining is not required. #3340 on paraffin-embedded HBL435 xenograft. Immunoprecipita |
| Immunogen Gel-purified p53 containing aa 14-389 of human p53. |
| Size and concentration 100µg and lot specific |
| Form liquid |
| Storage Instructions This antibody is stable for at least one (1) year at -20°C. Avoid multiple freeze-thaw cycles. |
| Storage buffer PBS, pH 7.4 |
| Purity protein affinity purification |
| Clonality monoclonal |
| Isotype IgG1 |
| Compatible secondaries goat anti-mouse IgG, H&L chain specific, peroxidase conjugated polyclonal antibody 5486 goat anti-mouse IgG, H&L chain specific, biotin conjugated, Conjugate polyclonal antibody 2685 goat anti-mouse IgG, H&L chain specific, FITC conjugated polyclonal antibody 7854 goat anti-mouse IgG, H&L chain specific, peroxidase conjugated polyclonal antibody, crossabsorbed 1706 goat anti-mouse IgG, H&L chain specific, biotin conjugated polyclonal antibody, crossabsorbed 1716 goat anti-mouse IgG, H&L chain specific, FITC conjugated polyclonal antibody, crossabsorbed 1721 |
| Isotype control Mouse monocolonal IgG1 - Isotype Control |
| target relevance |
|---|
| Protein names Cellular tumor antigen p53 (Antigen NY-CO-13) (Phosphoprotein p53) (Tumor suppressor p53) |
| Gene names TP53,TP53 P53 |
| Mass 43653Da |
Data
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| 36636131 | 3D printing biocompatible materials with Multi Jet Fusion for bioreactor applications. | Balasankar Meera Priyadarshini, Wai Kay Kok, Vishwesh Dikshit, Shilun Feng, King Ho Holden Li, Yi Zhang | Int J Bioprint 9:623 |
| 36582449 | Genetically engineered probiotics as catalytic glucose depriver for tumor starvation therapy. | Penghao Ji, Bolin An, Zhongming Jie, Liping Wang, Shuwen Qiu, Changhao Ge, Qihui Wu, Jianlin Shi, Minfeng Huo | Mater Today Bio 18:100515 |
| 36578521 | METTL14 Regulates Intestine Cellular Senescence through m(6)A Modification of Lamin B Receptor. | Zizhen Zhang, Meng Xue, Jingyu Chen, Zhuo Wang, Fangyu Ju, Jiaojiao Ni, Jiawei Sun, Haoyue Wu, Huimei Zheng, Ziwei Lou, Yawen Zhang, Xiaohang Yang, Shujie Chen, Yongmei Xi, Liangjing Wang | Oxid Med Cell Longev 2022:9096436 |
| 36430911 | Transcriptome Analysis of Goat Mammary Gland Tissue Reveals the Adaptive Strategies and Molecular Mechanisms of Lactation and Involution. | Rong Xuan, Jianmin Wang, Xiaodong Zhao, Qing Li, Yanyan Wang, Shanfeng Du, Qingling Duan, Yanfei Guo, Zhibin Ji, Tianle Chao | Int J Mol Sci 23:N/A |
| 36376321 | Replication collisions induced by de-repressed S-phase transcription are connected with malignant transformation of adult stem cells. | Ting Zhang, Carsten Künne, Dong Ding, Stefan Günther, Xinyue Guo, Yonggang Zhou, Xuejun Yuan, Thomas Braun | Nat Commun 13:6907 |
Protocols
| relevant to this product |
|---|
| Western blot IHC ICC |
Documents
| # | SDS | Certificate | |
|---|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | |||
Only logged in customers who have purchased this product may leave a review.







Reviews
There are no reviews yet.