| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| host | mouse |
| isotype | IgG |
| clonality | monoclonal |
| concentration | 1 mg/mL |
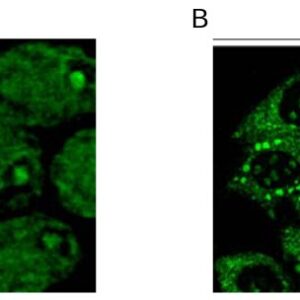
| applications | ICC/IF, WB |
| reactivity | Ago2 |
| available sizes | 200 µg |
mouse anti-Ago2 monoclonal antibody (1B1) 1524
$497.00
Antibody summary
- Mouse monoclonal to Ago2
- Suitable for: WB
- Isotype: Whole IgG
- 200 µg
mouse anti-Ago2 monoclonal antibody (1B1) 1524
| antibody |
|---|
| Tested applications WB |
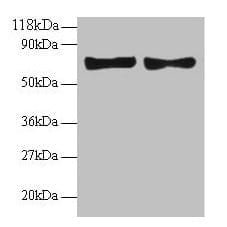
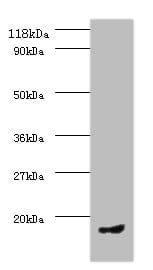
| Recommended dilutions Immunoblotting: use at 1-5ug/ml under reducing conditions. A band of ~100kDa is detected. These are recommended concentrations. End users should determine optimal concentrations for their applications. |
| Immunogen Recombinant protein corresponding to the N-terminal aa 1-148 of human Ago2. |
| Size and concentration 200µg and |
| Form lyophilized |
| Storage Instructions Store the stock solution at a minimum concentration of 2. 0mg/ml. A lower concentration may result in decreased stability. Store stock solution at -20°C for at least one (1) year; avoid mult |
| Storage buffer PBS, pH 7.4, 1.0% BSA, Lyophilized |
| Purity immunogen affinity purification |
| Clonality monoclonal |
| Isotype IgG2a |
| Compatible secondaries goat anti-mouse IgG, H&L chain specific, peroxidase conjugated polyclonal antibody 5486 goat anti-mouse IgG, H&L chain specific, biotin conjugated, Conjugate polyclonal antibody 2685 goat anti-mouse IgG, H&L chain specific, FITC conjugated polyclonal antibody 7854 goat anti-mouse IgG, H&L chain specific, peroxidase conjugated polyclonal antibody, crossabsorbed 1706 goat anti-mouse IgG, H&L chain specific, biotin conjugated polyclonal antibody, crossabsorbed 1716 goat anti-mouse IgG, H&L chain specific, FITC conjugated polyclonal antibody, crossabsorbed 1721 |
| Isotype control Mouse monocolonal IgG2a - Isotype Control |
| target relevance |
|---|
| Protein names Protein argonaute-2 (Argonaute2) (hAgo2) (EC 3.1.26.n2) (Argonaute RISC catalytic component 2) (Eukaryotic translation initiation factor 2C 2) (eIF-2C 2) (eIF2C 2) (PAZ Piwi domain protein) (PPD) (Protein slicer) |
| Gene names AGO2,AGO2 EIF2C2 |
| Protein family Argonaute family, Ago subfamily |
| Mass 97208Da |
| Function FUNCTION: Required for RNA-mediated gene silencing (RNAi) by the RNA-induced silencing complex (RISC). The 'minimal RISC' appears to include AGO2 bound to a short guide RNA such as a microRNA (miRNA) or short interfering RNA (siRNA). These guide RNAs direct RISC to complementary mRNAs that are targets for RISC-mediated gene silencing. The precise mechanism of gene silencing depends on the degree of complementarity between the miRNA or siRNA and its target. Binding of RISC to a perfectly complementary mRNA generally results in silencing due to endonucleolytic cleavage of the mRNA specifically by AGO2. Binding of RISC to a partially complementary mRNA results in silencing through inhibition of translation, and this is independent of endonuclease activity. May inhibit translation initiation by binding to the 7-methylguanosine cap, thereby preventing the recruitment of the translation initiation factor eIF4-E. May also inhibit translation initiation via interaction with EIF6, which itself binds to the 60S ribosomal subunit and prevents its association with the 40S ribosomal subunit. The inhibition of translational initiation leads to the accumulation of the affected mRNA in cytoplasmic processing bodies (P-bodies), where mRNA degradation may subsequently occur. In some cases RISC-mediated translational repression is also observed for miRNAs that perfectly match the 3' untranslated region (3'-UTR). Can also up-regulate the translation of specific mRNAs under certain growth conditions. Binds to the AU element of the 3'-UTR of the TNF (TNF-alpha) mRNA and up-regulates translation under conditions of serum starvation. Also required for transcriptional gene silencing (TGS), in which short RNAs known as antigene RNAs or agRNAs direct the transcriptional repression of complementary promoter regions. {ECO:0000250|UniProtKB:Q8CJG0, ECO:0000255|HAMAP-Rule:MF_03031, ECO:0000269|PubMed:15105377, ECO:0000269|PubMed:15260970, ECO:0000269|PubMed:15284456, ECO:0000269|PubMed:15337849, ECO:0000269|PubMed:15800637, ECO:0000269|PubMed:16081698, ECO:0000269|PubMed:16142218, ECO:0000269|PubMed:16271387, ECO:0000269|PubMed:16289642, ECO:0000269|PubMed:16357216, ECO:0000269|PubMed:16756390, ECO:0000269|PubMed:16936728, ECO:0000269|PubMed:17382880, ECO:0000269|PubMed:17507929, ECO:0000269|PubMed:17524464, ECO:0000269|PubMed:17531811, ECO:0000269|PubMed:17932509, ECO:0000269|PubMed:18048652, ECO:0000269|PubMed:18178619, ECO:0000269|PubMed:18690212, ECO:0000269|PubMed:18771919, ECO:0000269|PubMed:19167051, ECO:0000269|PubMed:23746446, ECO:0000269|PubMed:37328606}.; FUNCTION: (Microbial infection) Upon Sars-CoV-2 infection, associates with viral miRNA-like small RNA, CoV2-miR-O7a, and may repress mRNAs, such as BATF2, to evade the IFN response. {ECO:0000269|PubMed:34903581}. |
| Catalytic activity CATALYTIC ACTIVITY: Reaction=Endonucleolytic cleavage to 5'-phosphomonoester.; EC=3.1.26.n2; Evidence={ECO:0000255|HAMAP-Rule:MF_03031, ECO:0000269|PubMed:15105377, ECO:0000269|PubMed:23746446}; |
| Subellular location SUBCELLULAR LOCATION: Cytoplasm, P-body {ECO:0000269|PubMed:23125361, ECO:0000269|PubMed:23603119, ECO:0000269|PubMed:31400113, ECO:0000269|PubMed:33199684}. Nucleus {ECO:0000269|PubMed:23125361}. Note=Translational repression of mRNAs results in their recruitment to P-bodies. Translocation to the nucleus requires IMP8. |
| Structure SUBUNIT: Interacts with DICER1 through its Piwi domain and with TARBP2 during assembly of the RNA-induced silencing complex (RISC) (PubMed:14749716, PubMed:15973356, PubMed:16271387, PubMed:16289642, PubMed:16357216, PubMed:17507929, PubMed:18178619, PubMed:18690212, PubMed:33199684). Together, DICER1, AGO2 and TARBP2 constitute the trimeric RISC loading complex (RLC), or micro-RNA (miRNA) loading complex (miRLC). Within the RLC/miRLC, DICER1 and TARBP2 are required to process precursor miRNAs (pre-miRNAs) to mature miRNAs and then load them onto AGO2. AGO2 bound to the mature miRNA constitutes the minimal RISC and may subsequently dissociate from DICER1 and TARBP2. Note however that the term RISC has also been used to describe the trimeric RLC/miRLC. The formation of RISC complexes containing siRNAs rather than miRNAs appears to occur independently of DICER1. Interacts with AGO1. Also interacts with DDB1, DDX5, DDX6, DDX20, DHX30, DHX36, DDX47, DHX9, ELAVL, FXR1, GEMIN4, HNRNPF, IGF2BP1, ILF3, IMP8, MATR3, PABPC1, PRMT5, P4HA1, P4HB, RBM4, SART3, TNRC6A, TNRC6B, UPF1 and YBX1. Interacts with the P-body components DCP1A and XRN1. Associates with polysomes and messenger ribonucleoproteins (mNRPs). Interacts with RBM4; the interaction is modulated under stress-induced conditions, occurs under both cell proliferation and differentiation conditions and in an RNA- and phosphorylation-independent manner. Interacts with LIMD1, WTIP and AJUBA. Interacts with TRIM71; the interaction increases in presence of RNA (PubMed:23125361). Interacts with APOBEC3G in an RNA-dependent manner. Interacts with APOBEC3A, APOBEC3C, APOBEC3F and APOBEC3H. Interacts with DICER1, TARBP2, EIF6, MOV10 and RPL7A (60S ribosome subunit); they form a large RNA-induced silencing complex (RISC) (PubMed:17507929, PubMed:24726324). Interacts with FMR1 (PubMed:14703574). Interacts with ZFP36 (PubMed:15766526). Found in a complex, composed of AGO2, CHD7 and ARB2A (By similarity). Interacts with RC3H1; the interaction is RNA independent (PubMed:25697406). Interacts with SND1 (PubMed:14508492, PubMed:28546213). Interacts with SYT11 (By similarity). Interacts with CLNK (PubMed:26009488). Interacts with GARRE1 (PubMed:29395067). Interacts with GRB2; this interaction is important for the formation of a ternary complex containing GRB2, AGO2 and DICER1 (PubMed:37328606). {ECO:0000250|UniProtKB:Q8CJG0, ECO:0000255|HAMAP-Rule:MF_03031, ECO:0000269|PubMed:14508492, ECO:0000269|PubMed:14703574, ECO:0000269|PubMed:14749716, ECO:0000269|PubMed:15337849, ECO:0000269|PubMed:15766526, ECO:0000269|PubMed:15973356, ECO:0000269|PubMed:16081698, ECO:0000269|PubMed:16271387, ECO:0000269|PubMed:16289642, ECO:0000269|PubMed:16357216, ECO:0000269|PubMed:16699599, ECO:0000269|PubMed:16756390, ECO:0000269|PubMed:17382880, ECO:0000269|PubMed:17507929, ECO:0000269|PubMed:17531811, ECO:0000269|PubMed:17932509, ECO:0000269|PubMed:18178619, ECO:0000269|PubMed:18690212, ECO:0000269|PubMed:19167051, ECO:0000269|PubMed:19801630, ECO:0000269|PubMed:20505670, ECO:0000269|PubMed:20616046, ECO:0000269|PubMed:21475248, ECO:0000269|PubMed:21981923, ECO:0000269|PubMed:22539551, ECO:0000269|PubMed:22682761, ECO:0000269|PubMed:22791714, ECO:0000269|PubMed:22915799, ECO:0000269|PubMed:23125361, ECO:0000269|PubMed:24726324, ECO:0000269|PubMed:25697406, ECO:0000269|PubMed:26009488, ECO:0000269|PubMed:28546213, ECO:0000269|PubMed:29395067, ECO:0000269|PubMed:37328606}.; SUBUNIT: (Microbial infection) Interacts with Epstein-Barr virus (EBV) tegument protein BGLF2; this interaction participates in the regulation of cellular miRNA by the virus, leading to enhanced SUMOylation. {ECO:0000269|PubMed:32213613}.; SUBUNIT: (Microbial infection) Interacts with rotavirus A non-structural protein 5; this interaction probably plays a role in the sequestration of AGO2 in viral factories. {ECO:0000269|PubMed:30258011}.; SUBUNIT: (Microbial infection) Interacts with human herpesvirus 8 protein MTA/ORF57; this interaction inhibits P-body formation. {ECO:0000269|PubMed:31400113}. |
| Post-translational modification PTM: Hydroxylated. 4-hydroxylation appears to enhance protein stability but is not required for miRNA-binding or endonuclease activity. {ECO:0000255|HAMAP-Rule:MF_03031, ECO:0000269|PubMed:18690212}.; PTM: Ubiquitinated on surface-exposed lysines by a SCF-like E3 ubiquitin-protein ligase complex containing ZSWIM8 during target-directed microRNA degradation (TDMD), a process that mediates degradation of microRNAs (miRNAs) (PubMed:33184234, PubMed:33184237). Ubiquitination by the SCF-like E3 ubiquitin-protein ligase complex containing ZSWIM8 leads to its subsequent degradation, thereby exposing miRNAs for degradation (PubMed:33184234, PubMed:33184237). ZSWIM8 recognizes and binds AGO2 when it is engaged with a TDMD target (PubMed:33184237). {ECO:0000269|PubMed:33184234, ECO:0000269|PubMed:33184237}.; PTM: Phosphorylated. A phosphorylation cycle of C-terminal serine cluster (Ser-824-Ser-834) regulates the release of target mRNAs. Target-binding leads to phosphorylation of these residues by CSNK1A1, which reduces the affinity of AGO2 for mRNA and enables target release. The ANKRD52-PPP6C phosphatase complex dephosphorylates the residues, which primes AGO2 for binding a new target. {ECO:0000269|PubMed:28114302}.; PTM: Phosphorylation at Ser-387 by AKT3; leads to up-regulate translational repression of microRNA target and down-regulate endonucleolytic cleavage. {ECO:0000269|PubMed:23603119}. |
| Domain DOMAIN: The Piwi domain may perform RNA cleavage by a mechanism similar to that of RNase H. However, while RNase H utilizes a triad of Asp-Asp-Glu (DDE) for metal ion coordination, this protein appears to utilize a triad of Asp-Asp-His (DDH). |
| Involvement in disease DISEASE: Lessel-Kreienkamp syndrome (LESKRES) [MIM:619149]: An autosomal dominant disorder characterized by global developmental delay, intellectual disability of variable degree, and speech and language delay apparent from infancy or early childhood. Behavioral disorders are observed in most patients. Additional variable features include seizures, hypotonia, gait abnormalities, visual and cardiac defects, and non-specific facial dysmorphism. {ECO:0000269|PubMed:33199684}. Note=The disease is caused by variants affecting the gene represented in this entry. |
| Target Relevance information above includes information from UniProt accession: Q9UKV8 |
| The UniProt Consortium |
Data
| No results found |
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
| Western blot IHC ICC |
Documents
| # | SDS | Certificate | |
|---|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | |||
Only logged in customers who have purchased this product may leave a review.



Reviews
There are no reviews yet.