| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| accession | P0C6U8 |
| express system | E.coli |
| product tag | N-His-Avi |
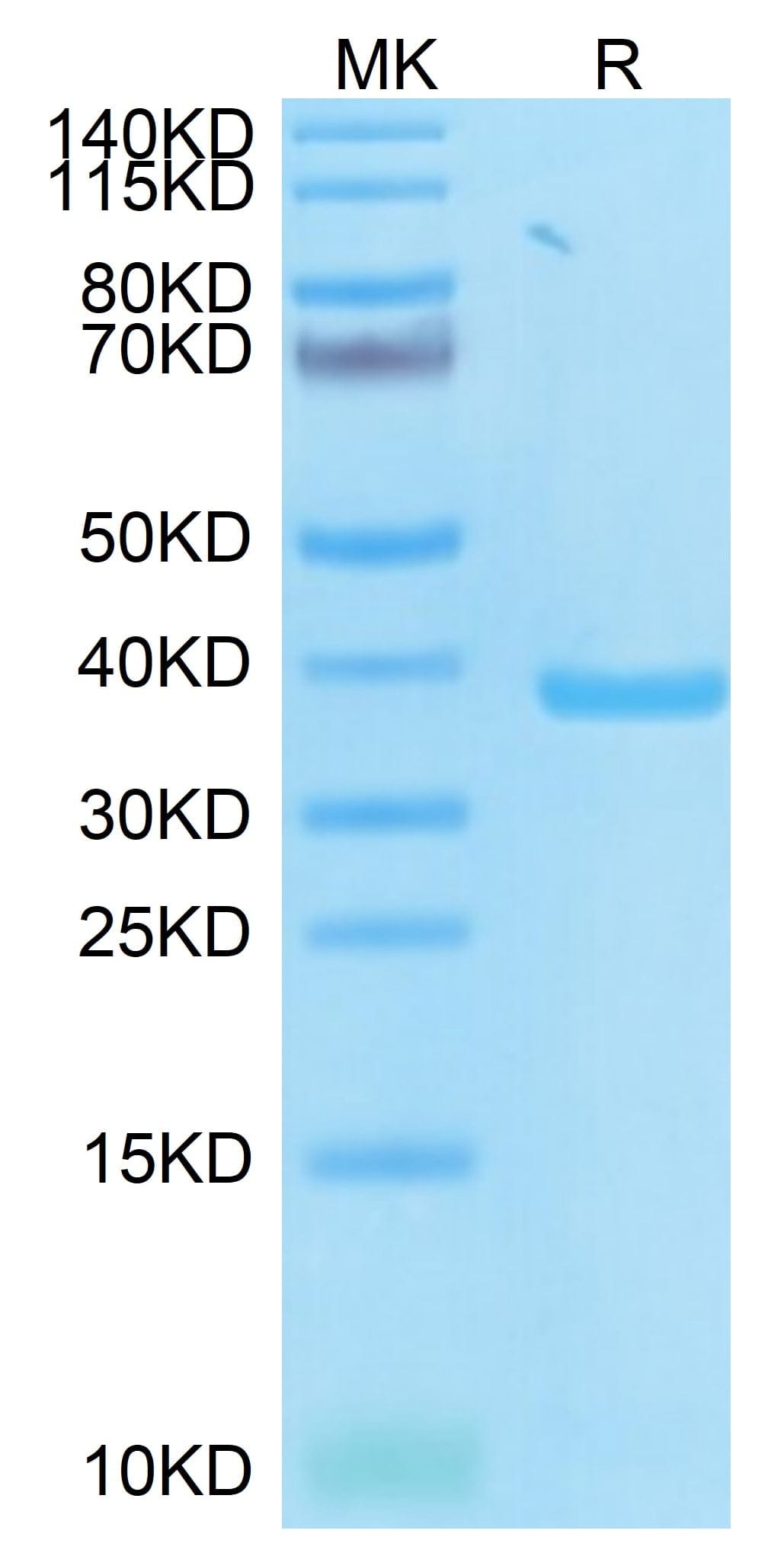
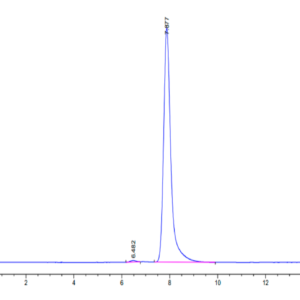
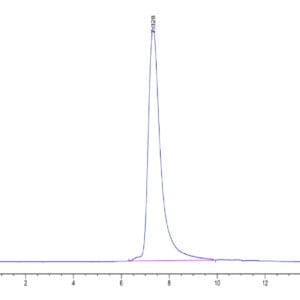
| purity | > 95% as determined by Tris-Bis PAGE |
| background | The coronaviral proteases, papain-like protease (PLpro) and 3C-like protease (3CLpro), are attractive antiviral drug targets because they are essential for coronaviral replication. Although the primary function of PLpro and 3CLpro are to process the viral polyprotein in a coordinated manner, PLpro has the additional function of stripping ubiquitin and ISG15 from host-cell proteins to aid coronaviruses in their evasion of the host innate immune responses. |
| molecular weight | The protein has a predicted MW of 38.6 kDa same as Tris-Bis PAGE result. |
| available size | 100 µg, 500 µg |
| endotoxin | Less than 1EU per μg by the LAL method. |
SARS PLpro/papain-like protease Protein 8372
$150.00 – $500.00
Summary
- Expression: E.coli
- Pure: Yes (SDS-PAGE)
- Amino Acid Range: Glu1-Ile314
SARS PLpro/papain-like protease Protein 8372
| protein |
|---|
| Size and concentration 100, 500µg and liquid |
| Form Liquid |
| Storage Instructions Valid for 12 months from date of receipt when stored at -80°C. Recommend to aliquot the protein into smaller quantities for optimal storage. Please minimize freeze-thaw cycles. |
| Storage buffer Shipped with dry ice. |
| Purity > 95% as determined by Tris-Bis PAGE |
| target relevance |
|---|
| The coronaviral proteases, papain-like protease (PLpro) and 3C-like protease (3CLpro), are attractive antiviral drug targets because they are essential for coronaviral replication. Although the primary function of PLpro and 3CLpro are to process the viral polyprotein in a coordinated manner, PLpro has the additional function of stripping ubiquitin and ISG15 from host-cell proteins to aid coronaviruses in their evasion of the host innate immune responses. |
| Protein names Replicase polyprotein 1a (pp1a) (ORF1a polyprotein) [Cleaved into: Host translation inhibitor nsp1 (Leader protein) (Non-structural protein 1) (nsp1); Non-structural protein 2 (nsp2) (p65 homolog); Papain-like protease nsp3 (PL-PRO) (EC 3.4.19.12) (EC 3.4.22.-) (Non-structural protein 3) (nsp3) (PL2-PRO); Non-structural protein 4 (nsp4); |
| Protein family Coronaviruses polyprotein 1ab family |
| Mass 486373Da |
| Function [Isoform Replicase polyprotein 1a]: Multifunctional protein involved in the transcription and replication of viral RNAs. Contains the proteinases responsible for the cleavages of the polyprotein. {ECO:0000305}.; [Host translation inhibitor nsp1]: Inhibits host translation by interacting with the 40S ribosomal subunit. The nsp1-40S ribosome complex further induces an endonucleolytic cleavage near the 5'UTR of host mRNAs, targeting them for degradation. Viral mRNAs are not susceptible to nsp1-mediated endonucleolytic RNA cleavage thanks to the presence of a 5'-end leader sequence and are therefore protected from degradation. By suppressing host gene expression, nsp1 facilitates efficient viral gene expression in infected cells and evasion from host immune response (PubMed:23035226). May disrupt nuclear pore function by binding and displacing host NUP93 (PubMed:30943371). {ECO:0000269|PubMed:23035226, ECO:0000269|PubMed:30943371}.; [Non-structural protein 2]: May play a role in the modulation of host cell survival signaling pathway by interacting with host PHB and PHB2 (PubMed:19640993). Indeed, these two proteins play a role in maintaining the functional integrity of the mitochondria and protecting cells from various stresses (PubMed:19640993). {ECO:0000269|PubMed:19640993}.; [Papain-like protease nsp3]: Responsible for the cleavages located at the N-terminus of the replicase polyprotein. In addition, PL-PRO possesses a deubiquitinating/deISGylating activity and processes both 'Lys-48'- and 'Lys-63'-linked polyubiquitin chains from cellular substrates (PubMed:17692280). Plays a role in host membrane rearrangement that leads to creation of cytoplasmic double-membrane vesicles (DMV) necessary for viral replication (PubMed:23943763). Nsp3, nsp4 and nsp6 together are sufficient to form DMV (PubMed:23943763, PubMed:24410069). Antagonizes innate immune induction of type I interferon by blocking the phosphorylation, dimerization and subsequent nuclear translocation of host IRF3 (PubMed:19369340, PubMed:24622840). Prevents also host NF-kappa-B signaling (PubMed:19369340, PubMed:24622840). {ECO:0000269|PubMed:16271890, ECO:0000269|PubMed:17692280, ECO:0000269|PubMed:19369340, ECO:0000269|PubMed:23943763, ECO:0000269|PubMed:24622840, ECO:0000303|PubMed:24410069}.; [Non-structural protein 4]: Plays a role in host membrane rearrangement that leads to creation of cytoplasmic double-membrane vesicles (DMV) necessary for viral replication (PubMed:23943763, PubMed:24410069). Alone appears incapable to induce membrane curvature, but together with nsp3 is able to induce paired membranes (PubMed:23943763). Nsp3, nsp4 and nsp6 together are sufficient to form DMV (PubMed:23943763, PubMed:24410069). {ECO:0000269|PubMed:23943763, ECO:0000303|PubMed:24410069}.; [3C-like proteinase nsp5]: Cleaves the C-terminus of replicase polyprotein at 11 sites. Recognizes substrates containing the core sequence [ILMVF]-Q-|-[SGACN]. Also able to bind an ADP-ribose-1''-phosphate (ADRP). May cleave host ATP6V1G1 thereby modifying host vacuoles intracellular pH. {ECO:0000255|PROSITE-ProRule:PRU00772, ECO:0000269|PubMed:16226257}.; [Non-structural protein 6]: Plays a role in host membrane rearrangement that leads to creation of cytoplasmic double-membrane vesicles (DMV) necessary for viral replication (PubMed:23943763). Nsp3, nsp4 and nsp6 together are sufficient to form DMV (PubMed:23943763, PubMed:24410069). Plays a role in the initial induction of autophagosomes from host reticulum endoplasmic. Later, limits the expansion of these phagosomes that are no longer able to deliver viral components to lysosomes (PubMed:24991833). {ECO:0000269|PubMed:23943763, ECO:0000269|PubMed:24991833, ECO:0000303|PubMed:24410069}.; [Non-structural protein 7]: Forms a hexadecamer with nsp8 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers. {ECO:0000269|PubMed:22039154}.; [Non-structural protein 8]: Forms a hexadecamer with nsp7 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers. {ECO:0000269|PubMed:22039154}.; [RNA-capping enzyme subunit nsp9]: Catalytic subunit of viral RNA capping enzyme which catalyzes the RNA guanylyltransferase reaction for genomic and sub-genomic RNAs. The kinase-like NiRAN domain of NSP12 transfers RNA to the amino terminus of NSP9, forming a covalent RNA-protein intermediate. Subsequently, the NiRAN domain transfers RNA to GDP, forming the core cap structure GpppA-RNA. The NSP14 and NSP16 methyltransferases then add methyl groups to form functional cap structures. {ECO:0000250|UniProtKB:P0DTC1, ECO:0000269|PubMed:19153232}.; [Non-structural protein 10]: Plays a pivotal role in viral transcription by stimulating both nsp14 3'-5' exoribonuclease and nsp16 2'-O-methyltransferase activities. Therefore plays an essential role in viral mRNAs cap methylation. {ECO:0000269|PubMed:22635272}. |
| Catalytic activity CATALYTIC ACTIVITY: [3C-like proteinase nsp5]: Reaction=TSAVLQ-|-SGFRK-NH2 and SGVTFQ-|-GKFKK the two peptides corresponding to the two self-cleavage sites of the SARS 3C-like proteinase are the two most reactive peptide substrates. The enzyme exhibits a strong preference for substrates containing Gln at P1 position and Leu at P2 position.; EC=3.4.22.69; Evidence={ECO:0000269|PubMed:12917450, ECO:0000269|PubMed:14561748}; CATALYTIC ACTIVITY: [Papain-like protease nsp3]: Reaction=Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).; EC=3.4.19.12; Evidence={ECO:0000269|PubMed:12917450, ECO:0000269|PubMed:17692280}; CATALYTIC ACTIVITY: [RNA-capping enzyme subunit nsp9]: Reaction=a 5'-end diphospho-ribonucleoside in mRNA + GTP + H(+) = a 5'-end (5'-triphosphoguanosine)-ribonucleoside in mRNA + diphosphate; Xref=Rhea:RHEA:67012, Rhea:RHEA-COMP:17165, Rhea:RHEA-COMP:17166, ChEBI:CHEBI:15378, ChEBI:CHEBI:33019, ChEBI:CHEBI:37565, ChEBI:CHEBI:167616, ChEBI:CHEBI:167617; EC=2.7.7.50; Evidence={ECO:0000250|UniProtKB:P0DTC1}; PhysiologicalDirection=right-to-left; Xref=Rhea:RHEA:67014; Evidence={ECO:0000250|UniProtKB:P0DTC1}; |
| Subellular location [Non-structural protein 2]: Host cytoplasm {ECO:0000250|UniProtKB:P0DTD1}. Host endosome {ECO:0000250|UniProtKB:P0DTD1}.; [Papain-like protease nsp3]: Host membrane {ECO:0000305}; Multi-pass membrane protein. Host cytoplasm {ECO:0000269|PubMed:23943763}.; [Non-structural protein 4]: Host membrane; Multi-pass membrane protein. Host cytoplasm {ECO:0000269|PubMed:23943763}. Note=Localizes in virally-induced cytoplasmic double-membrane vesicles. {ECO:0000269|PubMed:21345958, ECO:0000269|PubMed:23943763}.; [3C-like proteinase nsp5]: Host cytoplasm {ECO:0000250|UniProtKB:P0DTD1}. Host Golgi apparatus {ECO:0000250|UniProtKB:P0DTD1}.; [Non-structural protein 6]: Host membrane {ECO:0000305}; Multi-pass membrane protein {ECO:0000305}.; [Non-structural protein 7]: Host cytoplasm, host perinuclear region {ECO:0000250}. Host cytoplasm {ECO:0000250|UniProtKB:P0DTD1}. Host endoplasmic reticulum {ECO:0000250|UniProtKB:P0DTD1}. Note=nsp7, nsp8, nsp9 and nsp10 are localized in cytoplasmic foci, largely perinuclear. Late in infection, they merge into confluent complexes.; [Non-structural protein 8]: Host cytoplasm, host perinuclear region {ECO:0000269|PubMed:17532020}. Host cytoplasm {ECO:0000250|UniProtKB:P0DTD1}. Host endoplasmic reticulum {ECO:0000250|UniProtKB:P0DTD1}. Note=nsp7, nsp8, nsp9 and nsp10 are localized in cytoplasmic foci, largely perinuclear. Late in infection, they merge into confluent complexes.; [RNA-capping enzyme subunit nsp9]: Host cytoplasm, host perinuclear region {ECO:0000250}. Host cytoplasm {ECO:0000250|UniProtKB:P0DTD1}. Host endoplasmic reticulum {ECO:0000250|UniProtKB:P0DTD1}. Note=nsp7, nsp8, nsp9 and nsp10 are localized in cytoplasmic foci, largely perinuclear. Late in infection, they merge into confluent complexes.; [Non-structural protein 10]: Host cytoplasm, host perinuclear region {ECO:0000250}. Host cytoplasm {ECO:0000250|UniProtKB:P0DTD1}. Host endoplasmic reticulum {ECO:0000250|UniProtKB:P0DTD1}. Note=nsp7, nsp8, nsp9 and nsp10 are localized in cytoplasmic foci, largely perinuclear. Late in infection, they merge into confluent complexes. |
| Structure [Non-structural protein 2]: Interacts with host PHB and PHB2. {ECO:0000269|PubMed:19640993}.; [Non-structural protein 4]: Interacts with papain-like protease and non-structural protein 6. {ECO:0000269|PubMed:21345958}.; [3C-like proteinase nsp5]: Exists as monomer and homodimer. Only the homodimer shows catalytic activity. {ECO:0000269|PubMed:14561748, ECO:0000269|PubMed:15507456}.; [Non-structural protein 7]: Eight copies of nsp7 and eight copies of nsp8 assemble to form a heterohexadecamer dsRNA-encircling ring structure. {ECO:0000269|PubMed:16228002}.; [Non-structural protein 8]: Eight copies of nsp7 and eight copies of nsp8 assemble to form a heterohexadecamer dsRNA-encircling ring structure (PubMed:16228002). Interacts with ORF6 protein (PubMed:17532020). {ECO:0000269|PubMed:16228002, ECO:0000269|PubMed:17532020}.; [RNA-capping enzyme subunit nsp9]: Homodimer. {ECO:0000269|PubMed:19153232}.; [Non-structural protein 10]: Homododecamer. {ECO:0000269|PubMed:16873247}. |
| Post-translational modification [Isoform Replicase polyprotein 1a]: Specific enzymatic cleavages in vivo by its own proteases yield mature proteins (PubMed:12917450, PubMed:15331731, PubMed:15564471, PubMed:16306590, PubMed:32083638). 3C-like proteinase nsp5 liberates nsps 6-11 from the polyprotein (PubMed:32083638). Papain-like and 3C-like proteinases are autocatalytically processed. {ECO:0000269|PubMed:12917450, ECO:0000269|PubMed:15331731, ECO:0000269|PubMed:15564471, ECO:0000269|PubMed:16306590, ECO:0000269|PubMed:32083638}. |
| Domain [Papain-like protease nsp3]: The hydrophobic region HD1 probably mediates the membrane assoc |
| Target Relevance information above includes information from UniProt accession: P0C6U8 |
| The UniProt Consortium |
Publications
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
Documents
| # | ||
|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | ||