| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| accession | YP_009725299 |
| express system | E.coli |
| product tag | N-His |
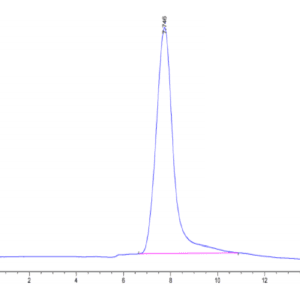
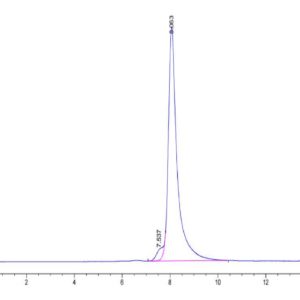
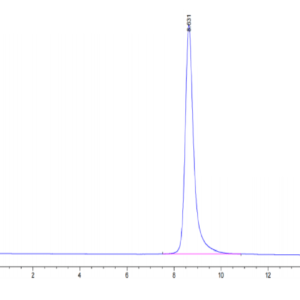
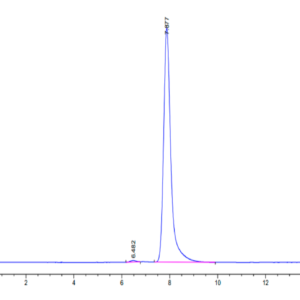
| purity | > 95% as determined by Tris-Bis PAGE |
| background | The coronaviral proteases, papain-like protease (PLpro) and 3C-like protease (3CLpro), are attractive antiviral drug targets because they are essential for coronaviral replication. Although the primary function of PLpro and 3CLpro are to process the viral polyprotein in a coordinated manner, PLpro has the additional function of stripping ubiquitin and ISG15 from host-cell proteins to aid coronaviruses in their evasion of the host innate immune responses. |
| molecular weight | The protein has a predicted MW of 39.2 kDa same as Tris-Bis PAGE result. |
| available size | 100 µg, 500 µg |
| endotoxin | Less than 1EU per μg by the LAL method. |
SARS-COV-2 PLpro/papain-like protease Protein 3219
$405.00 – $1,350.00
Summary
- Expression: E.coli
- Pure: Yes (SDS-PAGE)
- Amino Acid Range: Glu1564-Val1880
SARS-COV-2 PLpro/papain-like protease Protein 3219
| protein |
|---|
| https://www.uniprot.org/uniprotkb/P0DTC2/ |
| Size and concentration 100, 500µg and liquid |
| Form Liquid |
| Storage Instructions Valid for 12 months from date of receipt when stored at -80°C. Recommend to aliquot the protein into smaller quantities for optimal storage. Please minimize freeze-thaw cycles. |
| Storage buffer Shipped with dry ice. |
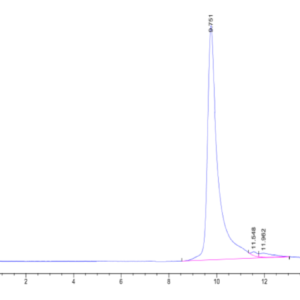
| Purity > 95% as determined by Tris-Bis PAGE |
| target relevance |
|---|
| The coronaviral proteases, papain-like protease (PLpro) and 3C-like protease (3CLpro), are attractive antiviral drug targets because they are essential for coronaviral replication. Although the primary function of PLpro and 3CLpro are to process the viral polyprotein in a coordinated manner, PLpro has the additional function of stripping ubiquitin and ISG15 from host-cell proteins to aid coronaviruses in their evasion of the host innate immune responses. |
| Protein names Spike glycoprotein (S glycoprotein) (E2) (Peplomer protein) [Cleaved into: Spike protein S1; Spike protein S2; Spike protein S2'] |
| Gene names S,S 2 |
| Protein family Betacoronaviruses spike protein family |
| Mass 141178Da |
| Function #N/A |
| Subellular location #N/A |
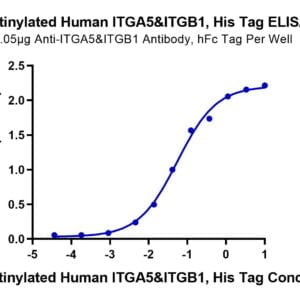
| Structure SUBUNIT: [Spike glycoprotein]: Homotrimer; each monomer consists of a S1 and a S2 subunit (PubMed:32075877, PubMed:32155444, PubMed:32245784). The resulting peplomers protrude from the virus surface as spikes (PubMed:32979942). Interacts with ORF3a protein and ORF7a protein (By similarity) (PubMed:32075877, PubMed:32155444, PubMed:32245784, PubMed:32979942). There are an average of 26 +/-15 spike trimers at the surface of virion particles (PubMed:32979942). Binds to host MBL2 (PubMed:35102342). This binding occurs via glycans and inhibits viral infectivity. Inhibition is effective against alpha, beta, gamma, and delta variants (PubMed:35102342). {ECO:0000255|HAMAP-Rule:MF_04099, ECO:0000269|PubMed:32075877, ECO:0000269|PubMed:32155444, ECO:0000269|PubMed:32245784, ECO:0000269|PubMed:32979942, ECO:0000269|PubMed:35102342}.; SUBUNIT: [Spike protein S1]: Binds to host ACE2 (PubMed:32075877, PubMed:32132184, PubMed:32155444, PubMed:32221306, PubMed:32225175, PubMed:32225176, PubMed:33607086). RBD also interacts with the N-linked glycan on 'Asn-90' of ACE2 (PubMed:33607086). Cleavage of S generates a polybasic C-terminal sequence on S1 that binds to host Neuropilin-1 (NRP1) and Neuropilin-2 (NRP2) receptors (PubMed:33082293, PubMed:33082294). Interacts with host integrin alpha-5/beta-1 (ITGA5:ITGB1) and with ACE2 in complex with integrin alpha-5/beta-1 (ITGA5:ITGB1) (PubMed:33102950). May interact via cytoplasmic c-terminus with M protein (PubMed:33229438). May interact (via N-terminus) with host bilirubin and biliverdin, thereby preventing antibody binding to the SARS-CoV-2 spike NTD via an allosteric mechanism (PubMed:33888467). May interact with host LRRC15, thereby allowing attachment to host cells (PubMed:36735681). {ECO:0000269|PubMed:32075877, ECO:0000269|PubMed:32132184, ECO:0000269|PubMed:32155444, ECO:0000269|PubMed:32221306, ECO:0000269|PubMed:32225175, ECO:0000269|PubMed:32225176, ECO:0000269|PubMed:33082293, ECO:0000269|PubMed:33082294, ECO:0000269|PubMed:33102950, ECO:0000269|PubMed:33229438, ECO:0000269|PubMed:33607086, ECO:0000269|PubMed:33888467, ECO:0000269|PubMed:36735681}. |
| Post-translational modification PTM: The cytoplasmic Cys-rich domain is palmitoylated. Palmitoylated spike proteins drive the formation of localized ordered cholesterol and sphingo-lipid-rich lipid nanodomains in the early Golgi, where viral budding occurs. {ECO:0000269|PubMed:34599882}.; PTM: Specific enzymatic cleavages in vivo yield mature proteins. The precursor is processed into S1 and S2 by host furin or unknown proteases to yield the mature S1 and S2 proteins (PubMed:32155444, PubMed:32362314, PubMed:32703818, PubMed:34159616, PubMed:34561887). Processing between S2 and S2' occurs either by host CTSL in endosomes (PubMed:32221306, PubMed:33465165, PubMed:34159616), or by host TMPRSS2 at the cell surface (PubMed:32142651). Both cleavages are necessary for the protein to be fusion competent (PubMed:32703818, PubMed:34159616, PubMed:34561887). Cell surface activation allows the virus to enter the cell despite inhibition of the endosomal pathway by hydroxychloroquine (PubMed:33465165). The polybasic furin cleavage site is absent in SARS-CoV S (PubMed:32155444, PubMed:32362314, PubMed:33465165). It increases the dependence on TMPRSS2 expression by SARS-CoV-2 (PubMed:33465165). D614G substitution would enhance furin cleavage at the S1/S2 junction (PubMed:33417835). {ECO:0000255|HAMAP-Rule:MF_04099, ECO:0000269|PubMed:32142651, ECO:0000269|PubMed:32155444, ECO:0000269|PubMed:32221306, ECO:0000269|PubMed:32362314, ECO:0000269|PubMed:32703818, ECO:0000269|PubMed:33417835, ECO:0000269|PubMed:33465165, ECO:0000269|PubMed:34159616, ECO:0000305|PubMed:34561887}.; PTM: Highly decorated by heterogeneous N-linked glycans protruding from the trimer surface (PubMed:32075877, PubMed:32155444, PubMed:32929138). Highly glycosylated by host both on S1 and S2 subunits, occluding many regions across the surface of the protein (PubMed:32363391, PubMed:32366695, PubMed:32929138). Approximately 40% of the protein surface is shielded from antibody recognition by glycans, with the notable exception of the ACE2 receptor binding domain (PubMed:32929138). {ECO:0000269|PubMed:32075877, ECO:0000269|PubMed:32155444, ECO:0000269|PubMed:32363391, ECO:0000269|PubMed:32366695, ECO:0000269|PubMed:32929138}.; PTM: O-glycosylated by host GALNT1 at the end of S1. This could reduce the efficiency of S1/S2 cleavage. {ECO:0000269|PubMed:34732583}. |
| Domain #N/A |
| Biotechnology BIOTECHNOLOGY: Main component of the anti-COVID19 vaccines BNT162b2/Pfizer-Biontech and mRNA-1273/Moderna; in which the mutations of Lys-986 (K986P) and Val-987 (V987P) have been added to stabilize the protein in the prefusion state. {ECO:0000305|PubMed:34322129}.; BIOTECHNOLOGY: Main component of the anti-COVID19 vaccine Ad26.COV2.S/Janssen Pharmaceutical; in which the mutations Arg-682 (R682S), Arg-685 (R685G), Lys-986 (K986P) and Val-987 (V987P) have been added to stabilize the protein in the prefusion state. {ECO:0000305|PubMed:34322129}.; BIOTECHNOLOGY: Main component of the anti-COVID vaccine Chadox1/AZD1222/AstraZeneca; in which the human tPA leader sequence is added in N-terminus to enhance protein secretion. {ECO:0000305|PubMed:34322129}. |
| Target Relevance information above includes information from UniProt accession: P0DTC2 |
| The UniProt Consortium |
Publications
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
Documents
| # | ||
|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | ||