| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| accession | O35305 |
| express system | HEK293 |
| product tag | C-hFc |
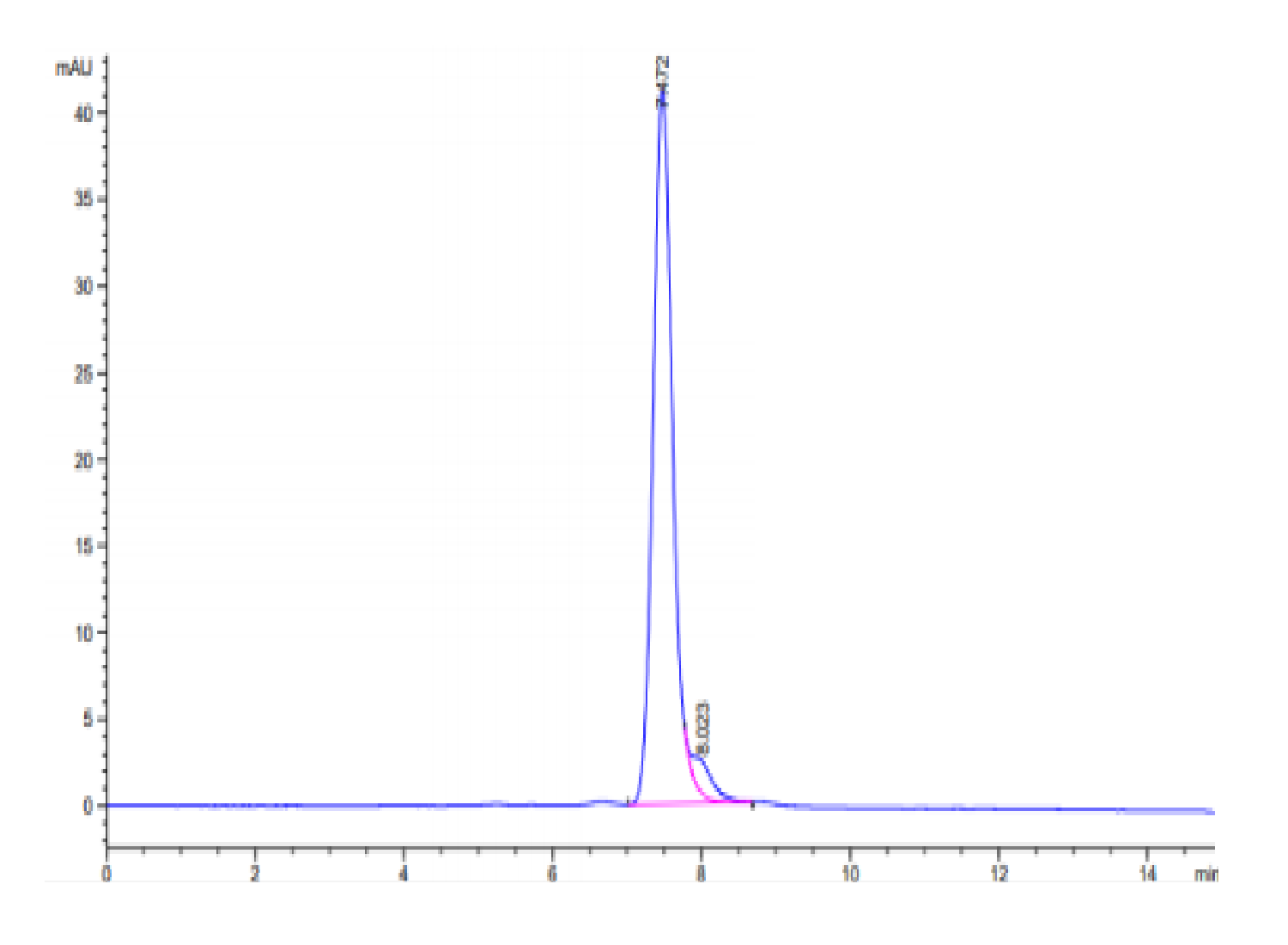
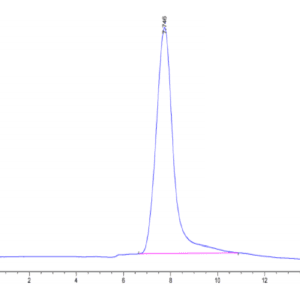
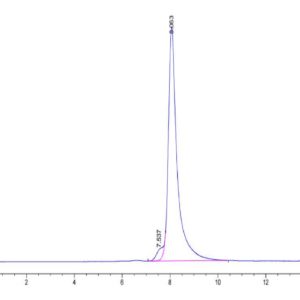
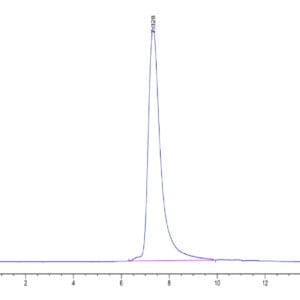
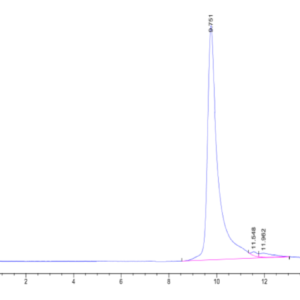
| purity | > 95% as determined by Tris-Bis PAGE;> 95% as determined by HPLC |
| background | TNFRSF11A, also known as receptor activator of NF-κB (RANK), activates several signaling pathways, such as NF-κB, JNK, ERK, p38α, and Akt/PKB. RANK/TNFRSF11A is a novel and frequent target for de novo methylation in gliomas, which affects apoptotic activity and focus formation thereby contributing to the molecular pathogenesis of gliomas. |
| molecular weight | The protein has a predicted MW of 47.0 kDa. Due to glycosylation, the protein migrates to 57-67 kDa based on Tris-Bis PAGE result. |
| available size | 100 µg, 500 µg |
| endotoxin | Less than 1EU per μg by the LAL method. |
Mouse TNFRSF11A/Rank Protein 2989
$315.00 – $1,050.00
Summary
- Expression: HEK293
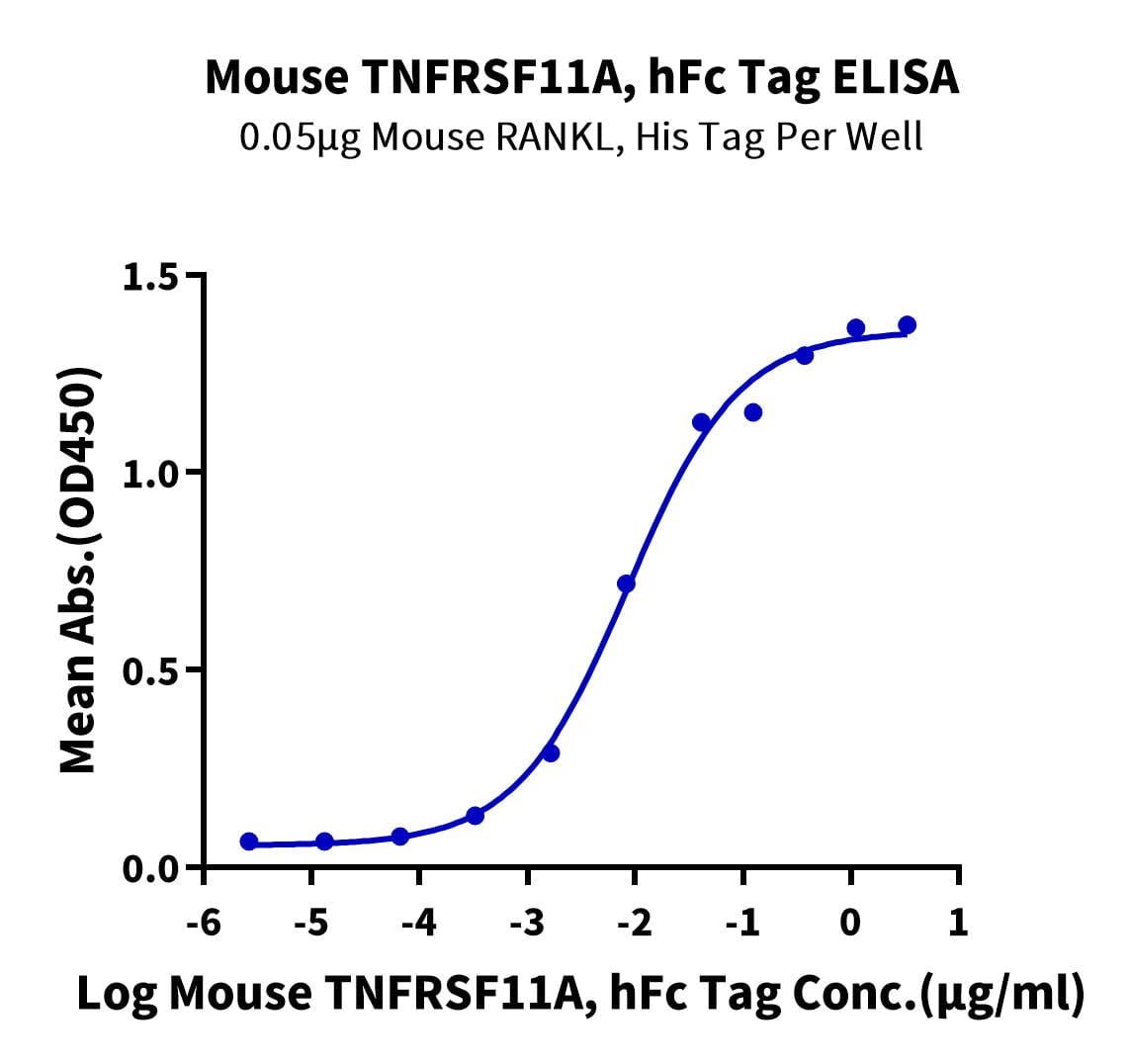
- Functional: Yes (ELISA)
- Amino Acid Range: Val31-Ser214
Mouse TNFRSF11A/Rank Protein 2989
| protein |
|---|
| Size and concentration 100, 500µg and lyophilized |
| Form Lyophilized |
| Storage Instructions Valid for 12 months from date of receipt when stored at -80°C. Recommend to aliquot the protein into smaller quantities for optimal storage. Please minimize freeze-thaw cycles. |
| Storage buffer Shipped at ambient temperature. |
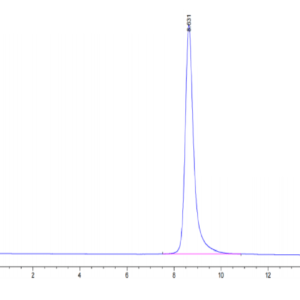
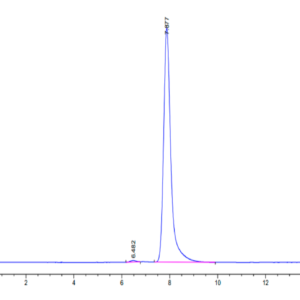
| Purity > 95% as determined by Tris-Bis PAGE |
| target relevance |
|---|
| TNFRSF11A, also known as receptor activator of NF-κB (RANK), activates several signaling pathways, such as NF-κB, JNK, ERK, p38α, and Akt/PKB. RANK/TNFRSF11A is a novel and frequent target for de novo methylation in gliomas, which affects apoptotic activity and focus formation thereby contributing to the molecular pathogenesis of gliomas. |
| Protein names Tumor necrosis factor receptor superfamily member 11A (Osteoclast differentiation factor receptor) (ODFR) (Receptor activator of NF-KB) (CD antigen CD265) |
| Gene names Tnfrsf11a,Tnfrsf11a Rank |
| Mass 10090Da |
| Function Receptor for TNFSF11/RANKL/TRANCE/OPGL; essential for RANKL-mediated osteoclastogenesis (PubMed:20483727, PubMed:23478294, PubMed:9878548). Its interaction with EEIG1 promotes osteoclastogenesis via facilitating the transcription of NFATC1 and activation of PLCG2 (PubMed:23478294). Involved in the regulation of interactions between T-cells and dendritic cells (PubMed:9367155). |
| Catalytic activity BINDING 134; /ligand="Na(+)"; /ligand_id="ChEBI:CHEBI:29101"; BINDING 135; /ligand="Na(+)"; /ligand_id="ChEBI:CHEBI:29101"; BINDING 138; /ligand="Na(+)"; /ligand_id="ChEBI:CHEBI:29101"; BINDING 161; /ligand="Na(+)"; /ligand_id="ChEBI:CHEBI:29101"; BINDING 163; /ligand="Na(+)"; /ligand_id="ChEBI:CHEBI:29101" |
| Subellular location Cell membrane ; Single-pass type I membrane protein. Membrane raft . |
| Tissues Ubiquitous expression with high levels in trabecular bone, thymus, small intestine, lung, brain and kidney. Weakly expressed in spleen and bone marrow. |
| Structure Binds to the clefts between the subunits of the TNFSF11 ligand trimer to form a heterohexamer (PubMed:20483727, PubMed:23039992). Part of a complex composed of EEIG1, TNFRSF11A/RANK, PLCG2, GAB2, TEC and BTK; complex formation increases in the presence of TNFSF11/RANKL (PubMed:23478294). Interacts with TRAF1, TRAF2, TRAF3, TRAF5 and TRAF6 (By similarity). Interacts (via cytoplasmic domain) with GAB2 (By similarity). Interacts (via cytoplasmic domain); with EEIG1 (via N-terminus); when in the presence of TNFSF11/RANKL (PubMed:23478294). |
| Target Relevance information above includes information from UniProt accession: O35305 |
| The UniProt Consortium |
Data
Publications
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
Documents
| # | ||
|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | ||