| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| accession | P56818 |
| express system | HEK293 |
| product tag | C-His |
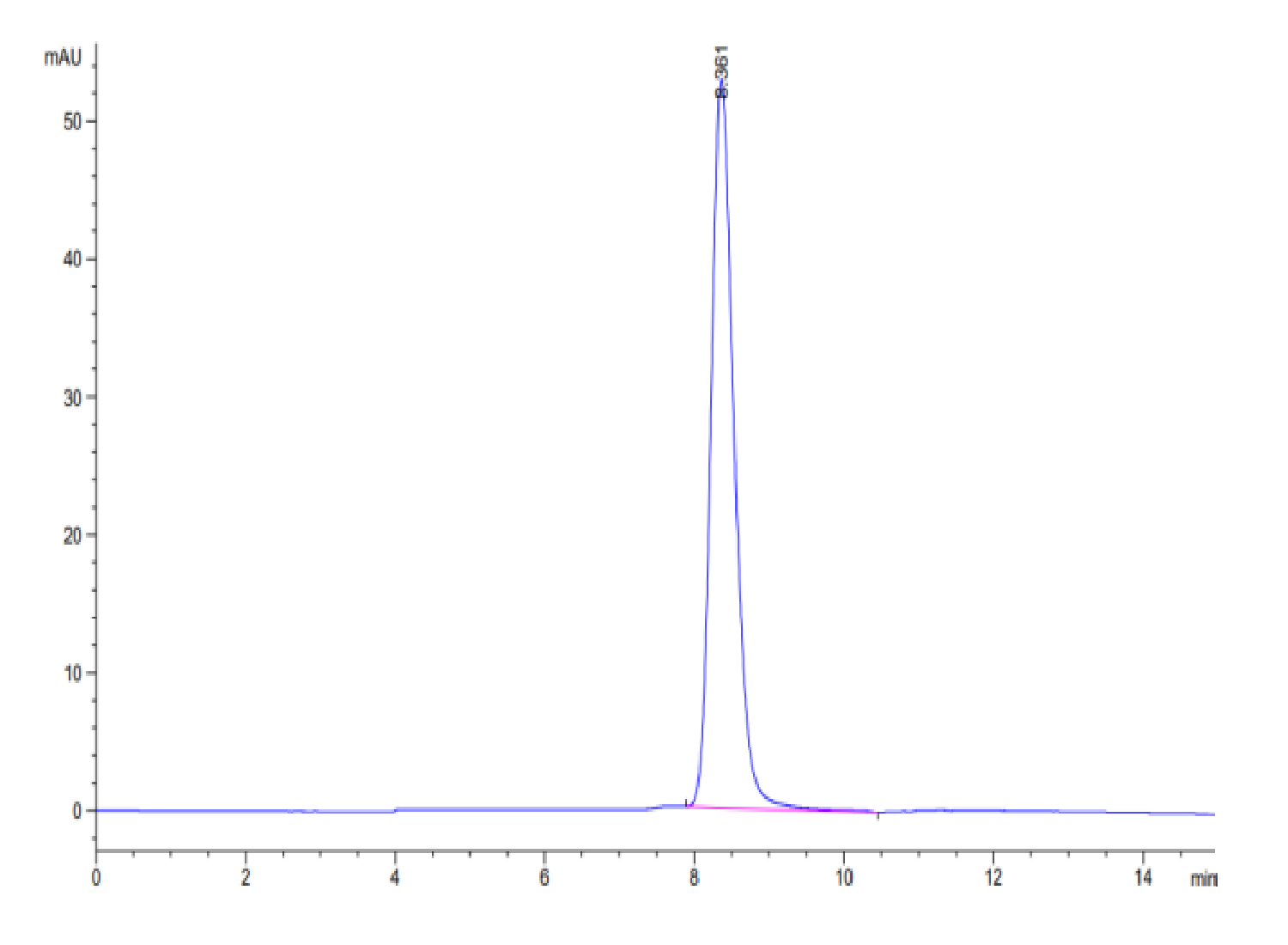
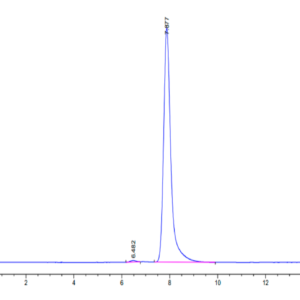
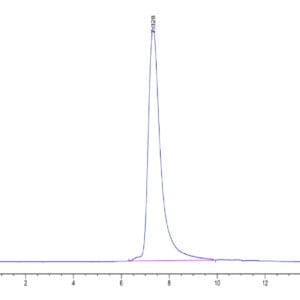
| purity | > 95% as determined by Tris-Bis PAGE;> 95% as determined by HPLC |
| background | The beta-site amyloid precursor protein cleaving enzyme-1 (BACE-1) initiates the generation of amyloid-β (Aβ), and the amyloid cascade leading to amyloid plaque deposition, neurodegeneration, and dementia in Alzheimer's disease (AD). Clinical failures of anti-Aβ therapies in dementia stages suggest that treatment has to start in the early, asymptomatic disease states. |
| molecular weight | The protein has a predicted MW of 49.49 kDa. Due to glycosylation, the protein migrates to 60-70 kDa based on Tris-Bis PAGE result. |
| available size | 100 µg, 500 µg |
| endotoxin | Less than 1EU per μg by the LAL method. |
Mouse BACE-1 Protein 3255
$240.00 – $800.00
Summary
- Expression: HEK293
- Active: Yes (catalytic)
- Amino Acid Range: Thr22-Thr457
Mouse BACE-1 Protein 3255
| protein |
|---|
| Size and concentration 100, 500µg and lyophilized |
| Form Lyophilized |
| Storage Instructions Valid for 12 months from date of receipt when stored at -80°C. Recommend to aliquot the protein into smaller quantities for optimal storage. Please minimize freeze-thaw cycles. |
| Storage buffer Shipped at ambient temperature. |
| Purity > 95% as determined by Tris-Bis PAGE |
| target relevance |
|---|
| The beta-site amyloid precursor protein cleaving enzyme-1 (BACE-1) initiates the generation of amyloid-β (Aβ), and the amyloid cascade leading to amyloid plaque deposition, neurodegeneration, and dementia in Alzheimer's disease (AD). Clinical failures of anti-Aβ therapies in dementia stages suggest that treatment has to start in the early, asymptomatic disease states. |
| Protein names Beta-secretase 1 (EC 3.4.23.46) (Aspartyl protease 2) (ASP2) (Asp 2) (Beta-site amyloid precursor protein cleaving enzyme 1) (Beta-site APP cleaving enzyme 1) (Memapsin-2) (Membrane-associated aspartic protease 2) |
| Gene names Bace1,Bace1 Bace |
| Protein family Peptidase A1 family |
| Mass 10090Da |
| Function Responsible for the proteolytic processing of the amyloid precursor protein (APP) (PubMed:29325091). Cleaves at the N-terminus of the A-beta peptide sequence, between residues 671 and 672 of APP, leads to the generation and extracellular release of beta-cleaved soluble APP, and a corresponding cell-associated C-terminal fragment which is later released by gamma-secretase (PubMed:29325091). Cleaves CHL1 (PubMed:29325091). |
| Subellular location Cell membrane ; Single-pass type I membrane protein. Golgi apparatus, trans-Golgi network. Endoplasmic reticulum. Endosome. Late endosome. Early endosome. Cell surface. Cytoplasmic vesicle membrane. Membrane raft. Lysosome. Recycling endosome. Cell projection, axon. Cell projection, dendrite. Note=Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (PubMed:25592972). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). |
| Tissues Expressed in the brain, specifically in neurons and astrocytes (at protein level). |
| Structure Monomer. Interacts (via DXXLL motif) with GGA1, GGA2 and GGA3 (via their VHS domain); the interaction highly increases when BACE1 is phosphorylated at Ser-498. Interacts with RTN1; RTN2; RTN3 and RTN4; the interaction leads to inhibition of amyloid precursor protein processing (By similarity). Interacts with SNX6. Interacts with PCSK9. Interacts with NAT8 and NAT8B. Interacts with BIN1 (By similarity). Interacts (via extracellular domain) with ADAM10 (via extracellular domain) (PubMed:29325091). Interacts with SORL1; this interaction may affect binding with APP and hence reduce APP cleavage (PubMed:16407538). Interacts with NRDC AND NRG1 (PubMed:19935654). |
| Post-translational modification N-Glycosylated (By similarity). Addition of a bisecting N-acetylglucosamine by MGAT3 blocks lysosomal targeting, further degradation and is required for maintaining stability under stress conditions (PubMed:25592972, PubMed:26467158).; Palmitoylation mediates lipid raft localization.; Acetylated in the endoplasmic reticulum at Lys-126, Lys-275, Lys-279, Lys-285, Lys-299, Lys-300 and Lys-307 (PubMed:20826464). Acetylation by NAT8 and NAT8B is transient and deacetylation probably occurs in the Golgi. Acetylation regulates the maturation, the transport to the plasma membrane, the stability and the expression of the protein.; Ubiquitinated at Lys-501, ubiquitination leads to lysosomal degradation. Monoubiquitinated and 'Lys-63'-linked polyubitinated. Deubiquitnated by USP8; inhibits lysosomal degradation.; Phosphorylation at Ser-498 is required for interaction with GGA1 and retrograded transport from endosomal compartments to the trans-Golgi network. Non-phosphorylated BACE1 enters a direct recycling route to the cell surface. |
| Domain DX |
| Target Relevance information above includes information from UniProt accession: P56818 |
| The UniProt Consortium |
Data
Publications
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
Documents
| # | ||
|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | ||