| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| accession | NP_085151 |
| express system | E.coli |
| product tag | N-His |
| purity | > 95% as determined by Tris-Bis PAGE;> 95% as determined by HPLC |
| background | SETD7 is a methyltransferase that specifically catalyzes the monomethylation of lysine 4 on histone H3. A variety of studies has revealed the role of SETD7 in posttranslational modifications of non-histone proteins. Aberrant expression of SETD7 has been associated with various diseases, including cancer. As a prognostic marker of breast cancer and a novel antioxidant promoter under oxidative stress in breast cancer, SETD7 is considered a good target for the development of new epigenetic drugs. |
| molecular weight | The protein has a predicted MW of 41.68 kDa. The protein migrates to 47-50 kDa based on Tris-Bis PAGE result. |
| available size | 100 ┬Ág, 500 ┬Ág |
| endotoxin | Less than 1EU per ╬╝g by the LAL method. |
Human SETD7 Protein 2467
$300.00 – $1,000.00
Summary
- Expression: E.coli
- Pure: Yes (HPLC)
- Amino Acid Range: Met1-Lys366
Human SETD7 Protein 2467
| protein |
|---|
| Size and concentration 100, 500µg and lyophilized |
| Form Lyophilized |
| Storage Instructions Valid for 12 months from date of receipt when stored at -80°C. Recommend to aliquot the protein into smaller quantities for optimal storage. Please minimize freeze-thaw cycles. |
| Storage buffer Shipped at ambient temperature. |
| Purity > 95% as determined by Tris-Bis PAGE |
| target relevance |
|---|
| SETD7 is a methyltransferase that specifically catalyzes the monomethylation of lysine 4 on histone H3. A variety of studies has revealed the role of SETD7 in posttranslational modifications of non-histone proteins. Aberrant expression of SETD7 has been associated with various diseases, including cancer. As a prognostic marker of breast cancer and a novel antioxidant promoter under oxidative stress in breast cancer, SETD7 is considered a good target for the development of new epigenetic drugs. |
| Protein names Histone-lysine N-methyltransferase SETD7 (EC 2.1.1.364) (Histone H3-K4 methyltransferase SETD7) (H3-K4-HMTase SETD7) (Lysine N-methyltransferase 7) (SET domain-containing protein 7) (SET7/9) |
| Protein family Class V-like SAM-binding methyltransferase superfamily, Histone-lysine methyltransf |
| Mass 40721Da |
| Function Histone methyltransferase that specifically monomethylates 'Lys-4' of histone H3 (PubMed:11779497, PubMed:11850410, PubMed:12540855, PubMed:12588998, PubMed:16141209). H3 'Lys-4' methylation represents a specific tag for epigenetic transcriptional activation (PubMed:12540855, PubMed:12588998, PubMed:16141209). Plays a central role in the transcriptional activation of genes such as collagenase or insulin (PubMed:12588998, PubMed:16141209). Recruited by IPF1/PDX-1 to the insulin promoter, leading to activate transcription (PubMed:16141209). Has also methyltransferase activity toward non-histone proteins such as CGAS, p53/TP53, TAF10, and possibly TAF7 by recognizing and binding the [KR]-[STA]-K in substrate proteins (PubMed:15099517, PubMed:15525938, PubMed:16415881, PubMed:35210392). Monomethylates 'Lys-189' of TAF10, leading to increase the affinity of TAF10 for RNA polymerase II (PubMed:15099517, PubMed:16415881). Monomethylates 'Lys-372' of p53/TP53, stabilizing p53/TP53 and increasing p53/TP53-mediated transcriptional activation (PubMed:15525938, PubMed:16415881, PubMed:17108971). Monomethylates 'Lys-491' of CGAS, promoting interaction between SGF29 and CGAS (By similarity). {ECO:0000250|UniProtKB:Q8VHL1, ECO:0000269|PubMed:11779497, ECO:0000269|PubMed:11850410, ECO:0000269|PubMed:12540855, ECO:0000269|PubMed:12588998, ECO:0000269|PubMed:15099517, ECO:0000269|PubMed:15525938, ECO:0000269|PubMed:16141209, ECO:0000269|PubMed:16415881, ECO:0000269|PubMed:17108971, ECO:0000269|PubMed:35210392}. |
| Catalytic activity CATALYTIC ACTIVITY: Reaction=L-lysyl(4)-[histone H3] + S-adenosyl-L-methionine = H(+) + N(6)-methyl-L-lysyl(4)-[histone H3] + S-adenosyl-L-homocysteine; Xref=Rhea:RHEA:60264, Rhea:RHEA-COMP:15543, Rhea:RHEA-COMP:15547, ChEBI:CHEBI:15378, ChEBI:CHEBI:29969, ChEBI:CHEBI:57856, ChEBI:CHEBI:59789, ChEBI:CHEBI:61929; EC=2.1.1.364; Evidence={ECO:0000255|PROSITE-ProRule:PRU00910, ECO:0000269|PubMed:12540855}; CATALYTIC ACTIVITY: Reaction=L-lysyl-[protein] + S-adenosyl-L-methionine = H(+) + N(6)-methyl-L-lysyl-[protein] + S-adenosyl-L-homocysteine; Xref=Rhea:RHEA:51736, Rhea:RHEA-COMP:9752, Rhea:RHEA-COMP:13053, ChEBI:CHEBI:15378, ChEBI:CHEBI:29969, ChEBI:CHEBI:57856, ChEBI:CHEBI:59789, ChEBI:CHEBI:61929; Evidence={ECO:0000269|PubMed:15099517, ECO:0000269|PubMed:15525938, ECO:0000269|PubMed:16415881}; PhysiologicalDirection=left-to-right; Xref=Rhea:RHEA:51737; Evidence={ECO:0000269|PubMed:15099517, ECO:0000269|PubMed:15525938, ECO:0000269|PubMed:16415881}; |
| Subellular location Nucleus {ECO:0000305|PubMed:11779497}. Chromosome {ECO:0000305|PubMed:11850410}. |
| Tissues Widely expressed. Expressed in pancreatic islets. {ECO:0000269|PubMed:16141209}. |
| Structure Interacts with IPF1/PDX-1. {ECO:0000269|PubMed:12514135, ECO:0000269|PubMed:12540855, ECO:0000269|PubMed:16141209}. |
| Domain The SET domain is necessary but not sufficient for histone methyltransferase activity. {ECO: |
| Target Relevance information above includes information from UniProt accession: Q8WTS6 |
| The UniProt Consortium |
Data
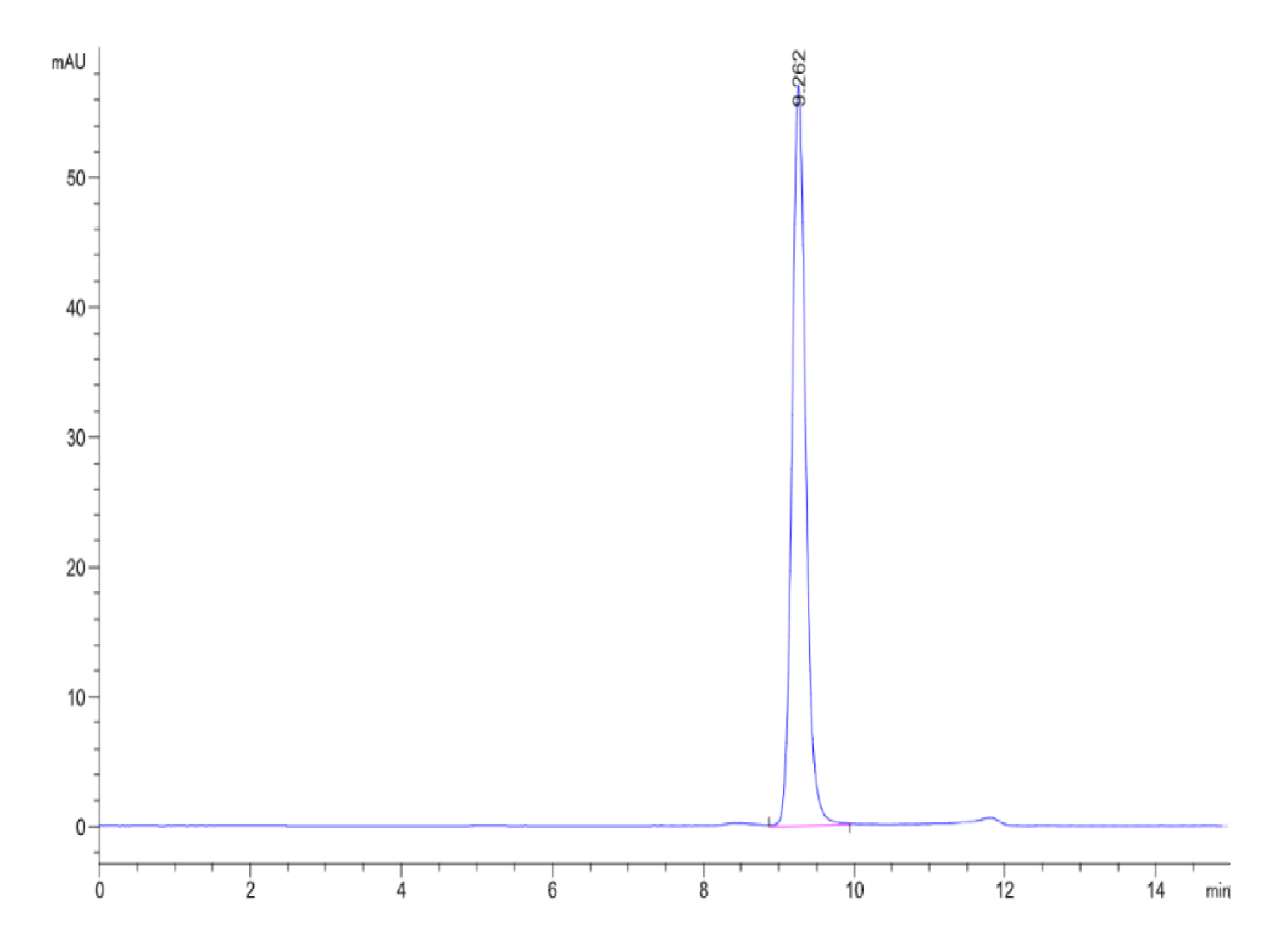 |
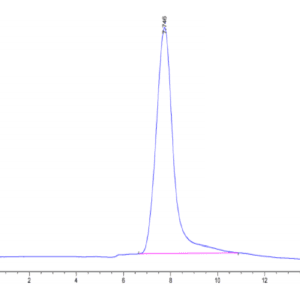
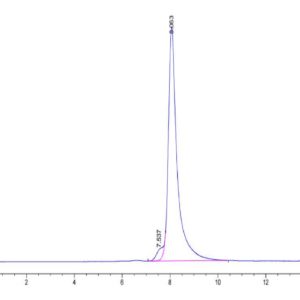
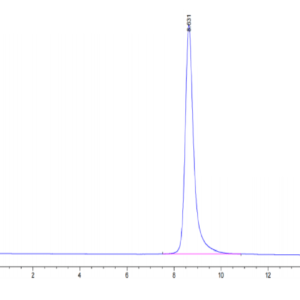
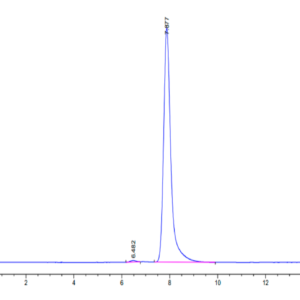
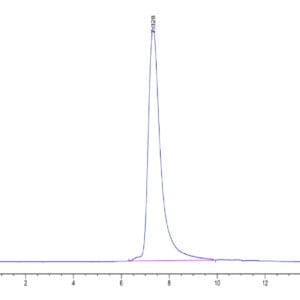
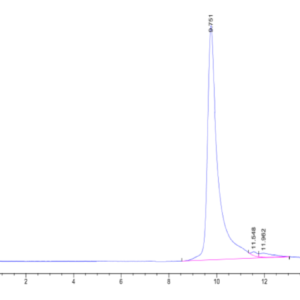
| The purity of Human SETD7 is greater than 95% as determined by SEC-HPLC. |
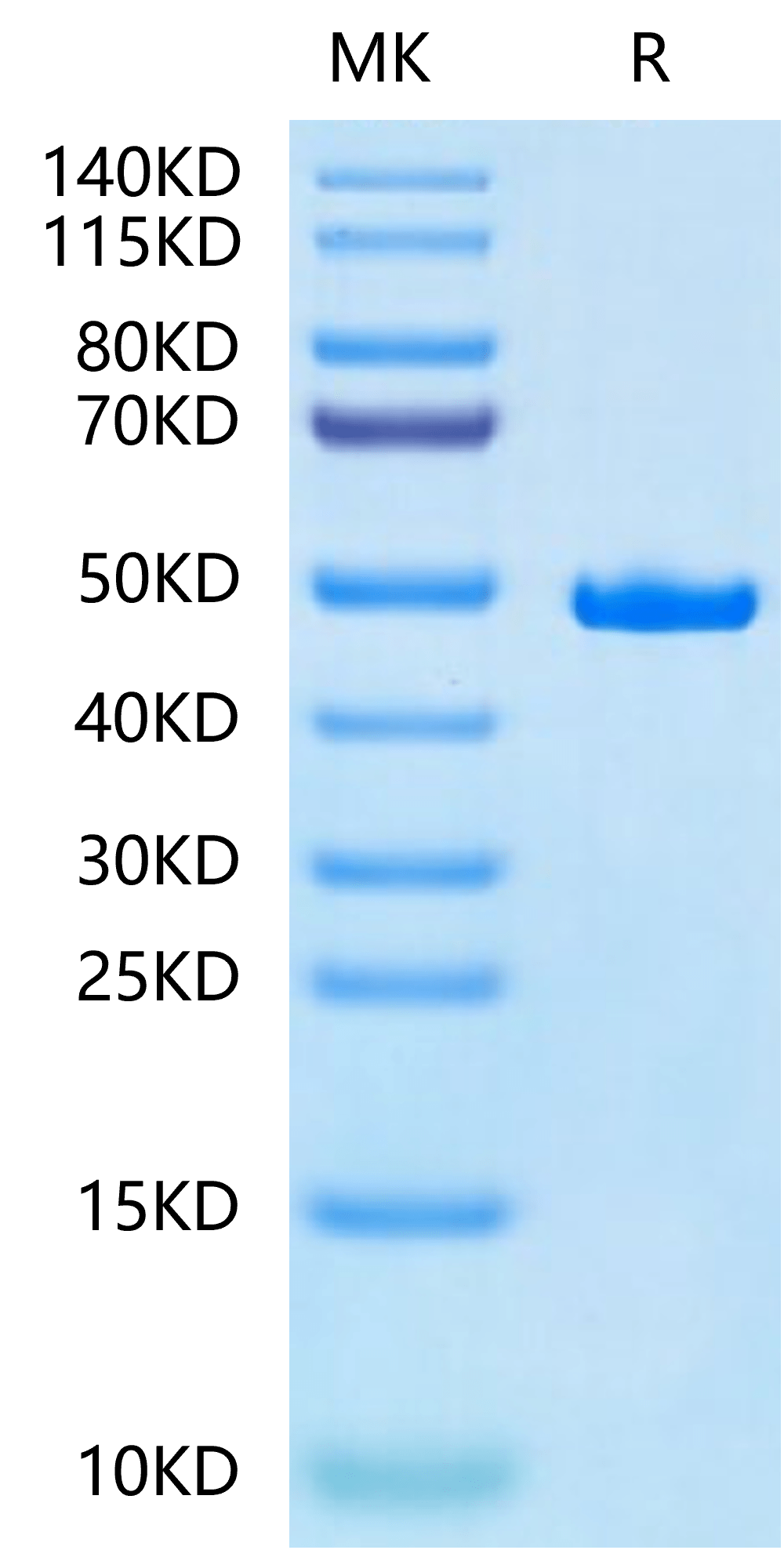 |
| Human SETD7 on Tris-Bis PAGE under reduced condition. The purity is greater than 95%. |
Publications
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
Documents
| # | ||
|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | ||