| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| accession | P01111 |
| express system | E.coli |
| product tag | N-His |
| purity | > 95% as determined by Tris-Bis PAGE;> 90% as determined by HPLC |
| background | NRas is a key mediator of the mitogenic pathway in normal cells and in cancer cells. Its dynamics and nanoscale organization at the plasma membrane (PM) facilitate its signaling. NRas activity is mediated and regulated by its dynamic interactions with functional (Grb2, BRAF, NF1, and GPI-nucleated) clusters at the PM. |
| molecular weight | The protein has a predicted MW of 22.29 kDa same as Tris-Bis PAGE result. |
| available size | 100 Āµg, 500 Āµg |
| endotoxin | Less than 1EU per Ī¼g by the LAL method. |
Human NRASĀ Protein 2432
$300.00 – $1,000.00
Summary
- Expression: E.coli
- Pure: Yes (HPLC)
- Amino Acid Range: Met1-Cys186
Human NRASĀ Protein 2432
| protein |
|---|
| Size and concentration 100, 500µg and lyophilized |
| Form Lyophilized |
| Storage Instructions Valid for 12 months from date of receipt when stored at -80°C. Recommend to aliquot the protein into smaller quantities for optimal storage. Please minimize freeze-thaw cycles. |
| Storage buffer Shipped at ambient temperature. |
| Purity > 95% as determined by Tris-Bis PAGE |
| target relevance |
|---|
| NRas is a key mediator of the mitogenic pathway in normal cells and in cancer cells. Its dynamics and nanoscale organization at the plasma membrane (PM) facilitate its signaling. NRas activity is mediated and regulated by its dynamic interactions with functional (Grb2, BRAF, NF1, and GPI-nucleated) clusters at the PM. |
| Protein names GTPase NRas (EC 3.6.5.2) (Transforming protein N-Ras) |
| Gene names NRAS,NRAS HRAS1 |
| Protein family Small GTPase superfamily, Ras family |
| Mass 9606Da |
| Function Ras proteins bind GDP/GTP and possess intrinsic GTPase activity. |
| Catalytic activity BINDING 10..18; /ligand="GTP"; /ligand_id="ChEBI:CHEBI:37565"; /evidence="ECO:0000269|Ref.25"; BINDING 29..30; /ligand="GTP"; /ligand_id="ChEBI:CHEBI:37565"; /evidence="ECO:0000269|Ref.25"; BINDING 57..61; /ligand="GTP"; /ligand_id="ChEBI:CHEBI:37565"; /evidence="ECO:0000255"; BINDING 116..119; /ligand="GTP"; /ligand_id="ChEBI:CHEBI:37565"; /evidence="ECO:0000269|Ref.25" |
| Subellular location Cell membrane ; Lipid-anchor ; Cytoplasmic side. Golgi apparatus membrane ; Lipid-anchor. Note=Shuttles between the plasma membrane and the Golgi apparatus. |
| Structure Interacts (active GTP-bound form preferentially) with RGS14 (By similarity). Interacts (active GTP-bound form) with RASSF7 (PubMed:21278800). |
| Post-translational modification Palmitoylated by the ZDHHC9-GOLGA7 complex (PubMed:16000296). Depalmitoylated by ABHD17A, ABHD17B and ABHD17C (PubMed:26701913). A continuous cycle of de- and re-palmitoylation regulates rapid exchange between plasma membrane and Golgi (PubMed:15705808, PubMed:16000296, PubMed:2661017, PubMed:26701913).; Acetylation at Lys-104 prevents interaction with guanine nucleotide exchange factors (GEFs).; Fatty-acylated at Lys-169 and/or Lys-170.; Ubiquitinated by the BCR(LZTR1) E3 ubiquitin ligase complex at Lys-170 in a non-degradative manner, leading to inhibit Ras signaling by decreasing Ras association with membranes.; Phosphorylation at Ser-89 by STK19 enhances NRAS-association with its downstream effectors.; (Microbial infection) Glucosylated at Thr-35 by P.sordellii toxin TcsL. |
| Target Relevance information above includes information from UniProt accession: P01111 |
| The UniProt Consortium |
Data
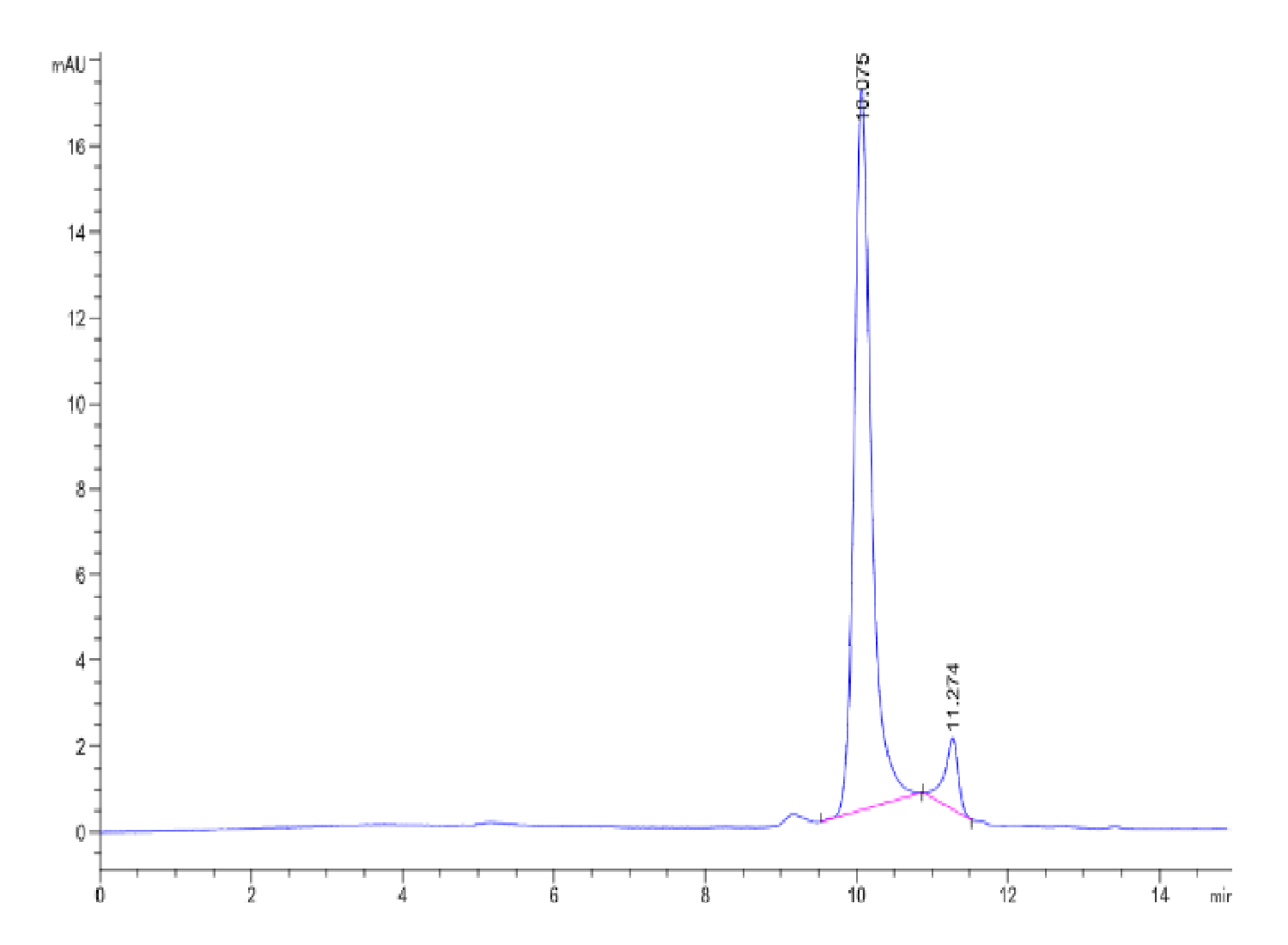 |
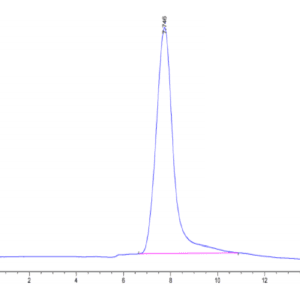
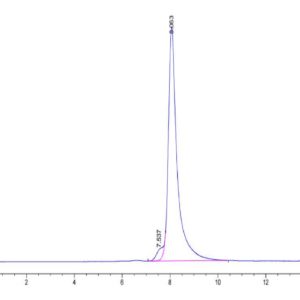
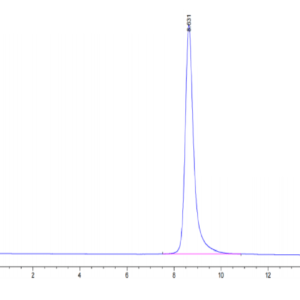
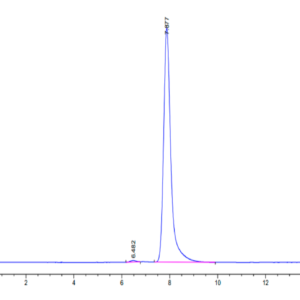
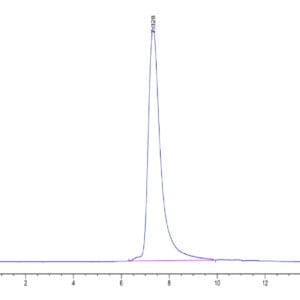
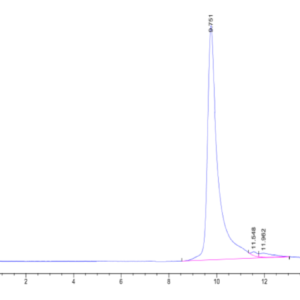
| The purity of Human NRAS is greater than 90% as determined by SEC-HPLC. |
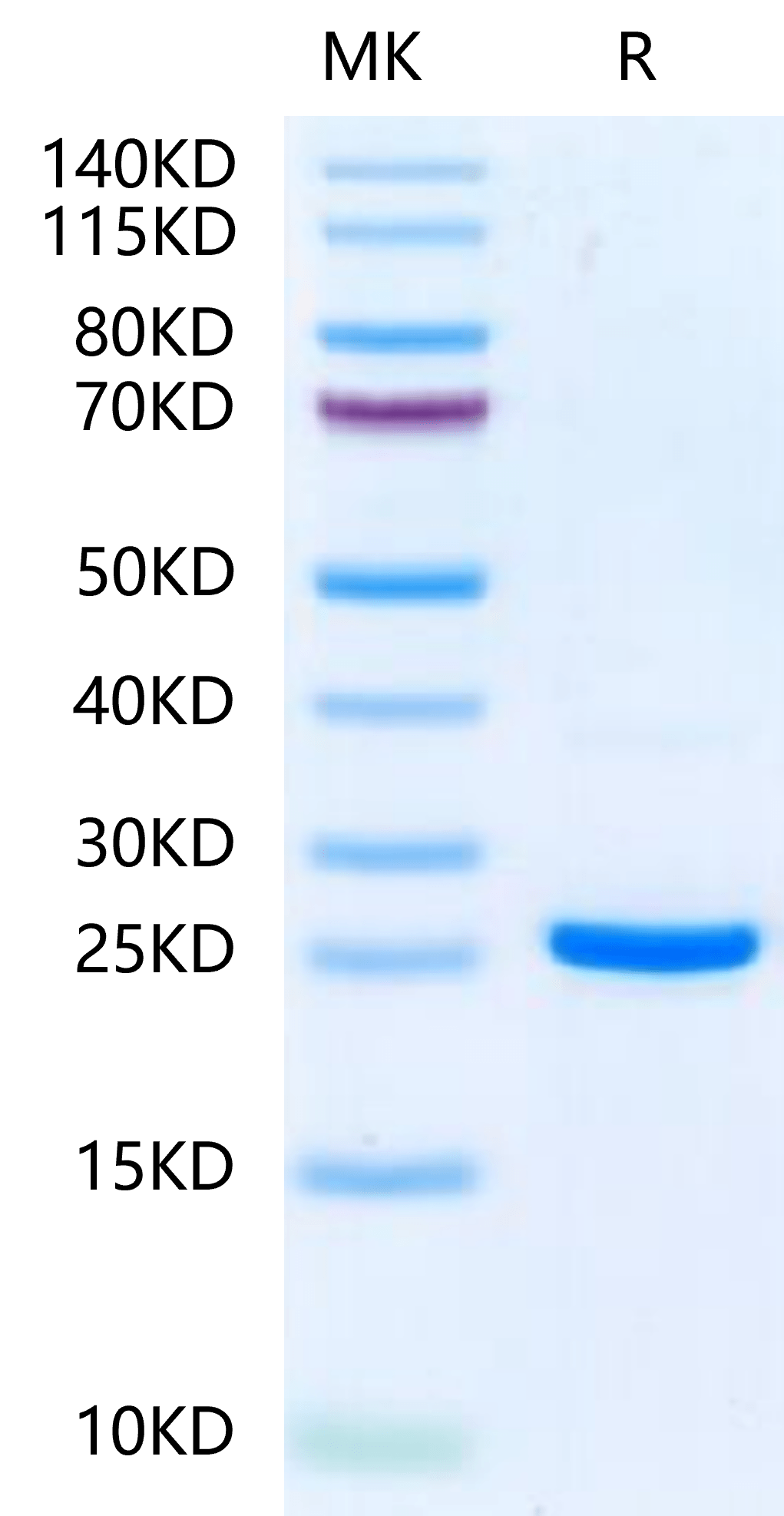 |
| Human NRAS on Tris-Bis PAGE under reduced condition. The purity is greater than 95%. |
Publications
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
Documents
| # | ||
|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | ||