| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| accession | YP_009725303(NSP7)&YP_009725304(NSP8) |
| express system | E.coli |
| product tag | C-His |
| purity | > 95% as determined by Tris-Bis PAGE |
| background | The crystal structure of the metabolite of remdesivir (Monophosphate of GS-441524) and NSP12-NSP8-NSP7 of SARS CoV-2 virus was recently reported. The crystal structures of ADP-Ribose or AMP and NSP3 of SARS CoV-2 virus were also released, recently. The crystal structure of NSP3 of SARS CoV-2 virus as an alternative binding site of AMP or ADP-ribose to treat COVID-19. |
| molecular weight | The protein has a predicted MW of 32.8 kDa same as Tris-Bis PAGE result. |
| available size | 100 µg, 500 µg |
| endotoxin | Less than 1EU per μg by the LAL method. |
SARS-COV-2 NSP7&NSP8 Protein 4364
$345.00 – $1,150.00
Summary
- Expression: E.coli
- Pure: Yes (SDS-PAGE)
- Amino Acid Range: Ser1-Gln83(NSP7) & Ala1-Gln198(NSP8)
SARS-COV-2 NSP7&NSP8 Protein 4364
| protein |
|---|
| https://www.uniprot.org/uniprotkb/P0DTC1/ |
| Size and concentration 100, 500µg and lyophilized |
| Form Lyophilized |
| Storage Instructions Valid for 12 months from date of receipt when stored at -80°C. Recommend to aliquot the protein into smaller quantities for optimal storage. Please minimize freeze-thaw cycles. |
| Storage buffer Shipped at ambient temperature. |
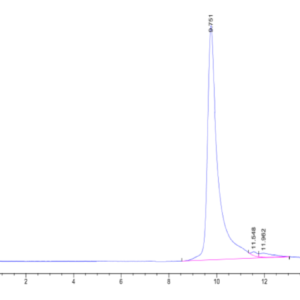
| Purity > 95% as determined by Tris-Bis PAGE |
| target relevance |
|---|
| The crystal structure of the metabolite of remdesivir (Monophosphate of GS-441524) and NSP12-NSP8-NSP7 of SARS CoV-2 virus was recently reported. The crystal structures of ADP-Ribose or AMP and NSP3 of SARS CoV-2 virus were also released, recently. The crystal structure of NSP3 of SARS CoV-2 virus as an alternative binding site of AMP or ADP-ribose to treat COVID-19. |
| Protein names Replicase polyprotein 1a (pp1a) (ORF1a polyprotein) [Cleaved into: Host translation inhibitor nsp1 (Leader protein) (Non-structural protein 1) (nsp1); Non-structural protein 2 (nsp2) (p65 homolog); Papain-like protease nsp3 (EC 3.4.19.12) (EC 3.4.22.-) (Non-structural protein 3) (nsp3) (PL2-PRO) (Papain-like proteinase) (PL-PRO); Non-stru |
| Protein family Coronaviruses polyprotein 1ab family |
| Mass 489989Da |
| Function #N/A |
| Catalytic activity CATALYTIC ACTIVITY: [Papain-like protease nsp3]: Reaction=Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).; EC=3.4.19.12; Evidence={ECO:0000269|PubMed:32726803}; CATALYTIC ACTIVITY: [3C-like proteinase nsp5]: Reaction=TSAVLQ-|-SGFRK-NH2 and SGVTFQ-|-GKFKK the two peptides corresponding to the two self-cleavage sites of the SARS 3C-like proteinase are the two most reactive peptide substrates. The enzyme exhibits a strong preference for substrates containing Gln at P1 position and Leu at P2 position.; EC=3.4.22.69; Evidence={ECO:0000269|PubMed:32198291, ECO:0000269|PubMed:32272481, ECO:0000269|PubMed:32321856}; CATALYTIC ACTIVITY: [RNA-capping enzyme subunit nsp9]: Reaction=a 5'-end diphospho-ribonucleoside in mRNA + GTP + H(+) = a 5'-end (5'-triphosphoguanosine)-ribonucleoside in mRNA + diphosphate; Xref=Rhea:RHEA:67012, Rhea:RHEA-COMP:17165, Rhea:RHEA-COMP:17166, ChEBI:CHEBI:15378, ChEBI:CHEBI:33019, ChEBI:CHEBI:37565, ChEBI:CHEBI:167616, ChEBI:CHEBI:167617; EC=2.7.7.50; Evidence={ECO:0000269|PubMed:35944563}; PhysiologicalDirection=right-to-left; Xref=Rhea:RHEA:67014; Evidence={ECO:0000269|PubMed:35944563}; |
| Subellular location #N/A |
| Structure SUBUNIT: [Non-structural protein 2]: Interacts with host PHB and PHB2. {ECO:0000250|UniProtKB:P0C6X7}.; SUBUNIT: [Papain-like protease nsp3]: May form homohexamers (PubMed:32763915). Interacts with N protein (PubMed:35044811). {ECO:0000269|PubMed:32763915, ECO:0000269|PubMed:35044811}.; SUBUNIT: [3C-like proteinase nsp5]: 3CL-PRO exists as monomer and homodimer. Only the homodimer shows catalytic activity. {ECO:0000269|PubMed:32198291}.; SUBUNIT: [Non-structural protein 4]: Interacts with PL-PRO and nsp6. {ECO:0000250|UniProtKB:P0C6X7}.; SUBUNIT: [Non-structural protein 6]: Forms homodimers (PubMed:35551511). Interacts with host ZFYVE1 (DFCP1) (PubMed:35551511), which leads to ER and DMVs binding to lipid droplets. Interacts with host TBK1; this interaction decreases IRF3 phosphorylation by 57%, which leads to reduced IFN-beta production. {ECO:0000269|PubMed:32979938, ECO:0000269|PubMed:35551511}.; SUBUNIT: [Non-structural protein 7]: Interacts with nsp8 and nsp12 to form the replication-transcription complex (RTC): nsp12, nsp7, two subunits of nsp8, and up to two subunits of nsp13 (PubMed:32277040, PubMed:32358203, PubMed:32438371, PubMed:32526208, PubMed:34562452). Eight copies of nsp7 and eight copies of nsp8 assemble to form a heterohexadecamer dsRNA-encircling ring structure (By similarity). {ECO:0000250|UniProtKB:P0C6X7, ECO:0000269|PubMed:32277040, ECO:0000269|PubMed:32358203, ECO:0000269|PubMed:32438371, ECO:0000269|PubMed:32526208, ECO:0000305|PubMed:34562452}.; SUBUNIT: [Non-structural protein 8]: Interacts with nsp7, nsp13 and nsp12 (PubMed:33232691) to form the replication-transcription complex (RTC): nsp12, nsp7, two subunits of nsp8, and up to two subunits of nsp13 (PubMed:32277040, PubMed:32358203, PubMed:32438371, PubMed:32526208, PubMed:34562452). Eight copies of nsp7 and eight copies of nsp8 assemble to form a heterohexadecamer dsRNA-encircling ring structure (By similarity). {ECO:0000250|UniProtKB:P0C6X7, ECO:0000269|PubMed:32277040, ECO:0000269|PubMed:32358203, ECO:0000269|PubMed:32438371, ECO:0000269|PubMed:32526208, ECO:0000269|PubMed:33232691, ECO:0000305|PubMed:34562452}.; SUBUNIT: [RNA-capping enzyme subunit nsp9]: Is a dimer (By similarity). Interacts with NSP12 (PubMed:35944563). {ECO:0000250|UniProtKB:P0C6X7, ECO:0000269|PubMed:35944563}.; SUBUNIT: [Non-structural protein 10]: Forms a dodecamer and interacts with nsp14 and nsp16; these interactions enhance nsp14 and nsp16 enzymatic activities. {ECO:0000250|UniProtKB:P0C6X7}. |
| Post-translational modification PTM: Specific enzymatic cleavages in vivo by its own proteases yield mature proteins. 3CL-PRO and PL-PRO proteinases are autocatalytically processed. {ECO:0000250|UniProtKB:P0C6X7}. |
| Domain #N/A |
| Target Relevance information above includes information from UniProt accession: P0DTC1 |
| The UniProt Consortium |
Publications
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
Documents
| # | ||
|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | ||