| Weight | 1 lbs |
|---|---|
| Dimensions | 9 × 5 × 2 in |
| accession | Q9GK78 |
| express system | HEK293 |
| product tag | C-His |
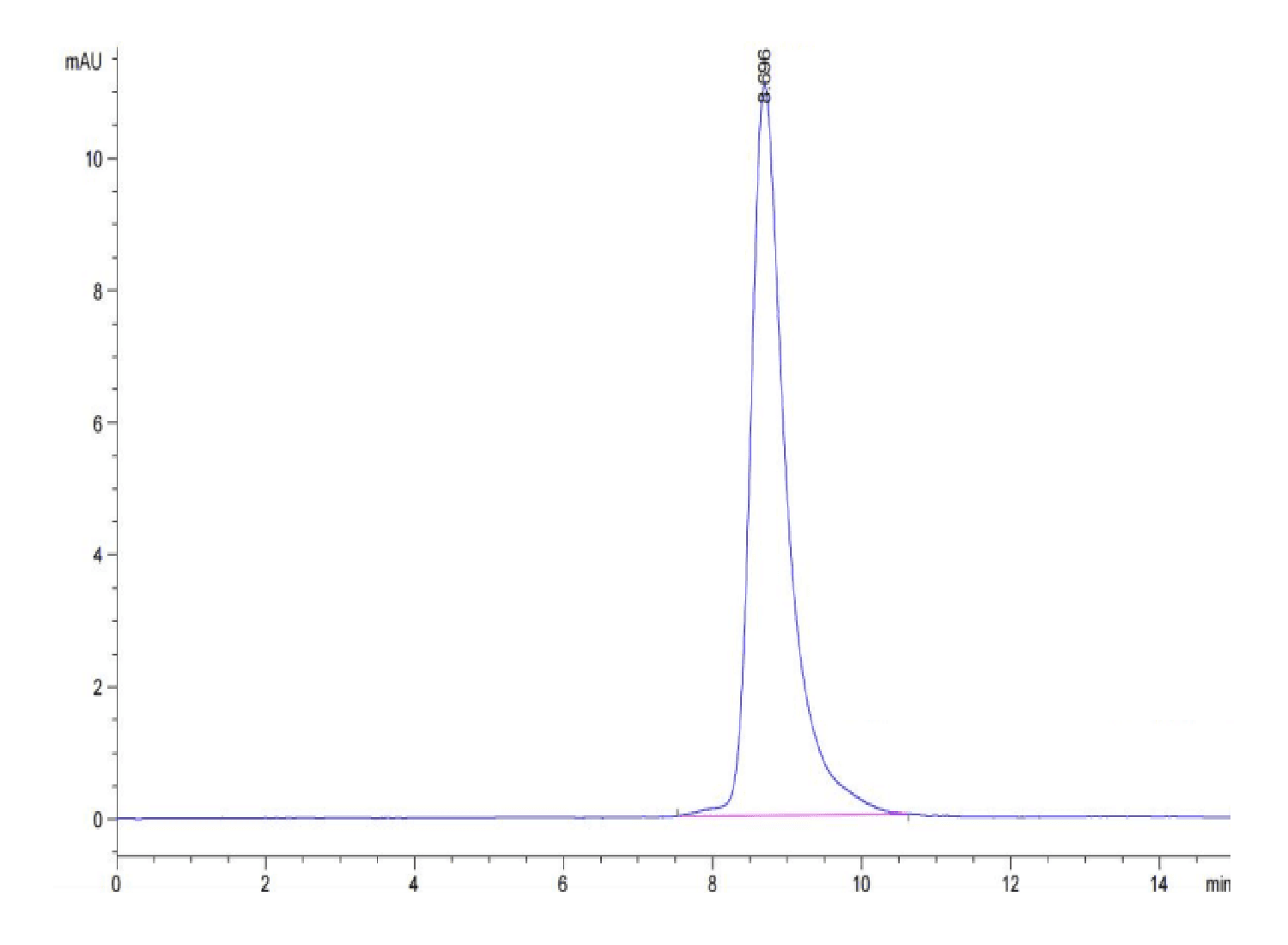
| purity | > 95% as determined by Tris-Bis PAGE;> 95% as determined by HPLC |
| background | The receptor (u-PAR) for urokinase plasminogen activator (u-PA) is a three-domain protein, GPI-anchored to the cell surface, which focuses the enzymatic activity of u-PA, and allows the cell surface activation of plasminogen.Regulation of the activity of u-PA is also mediated by u-PAR. |
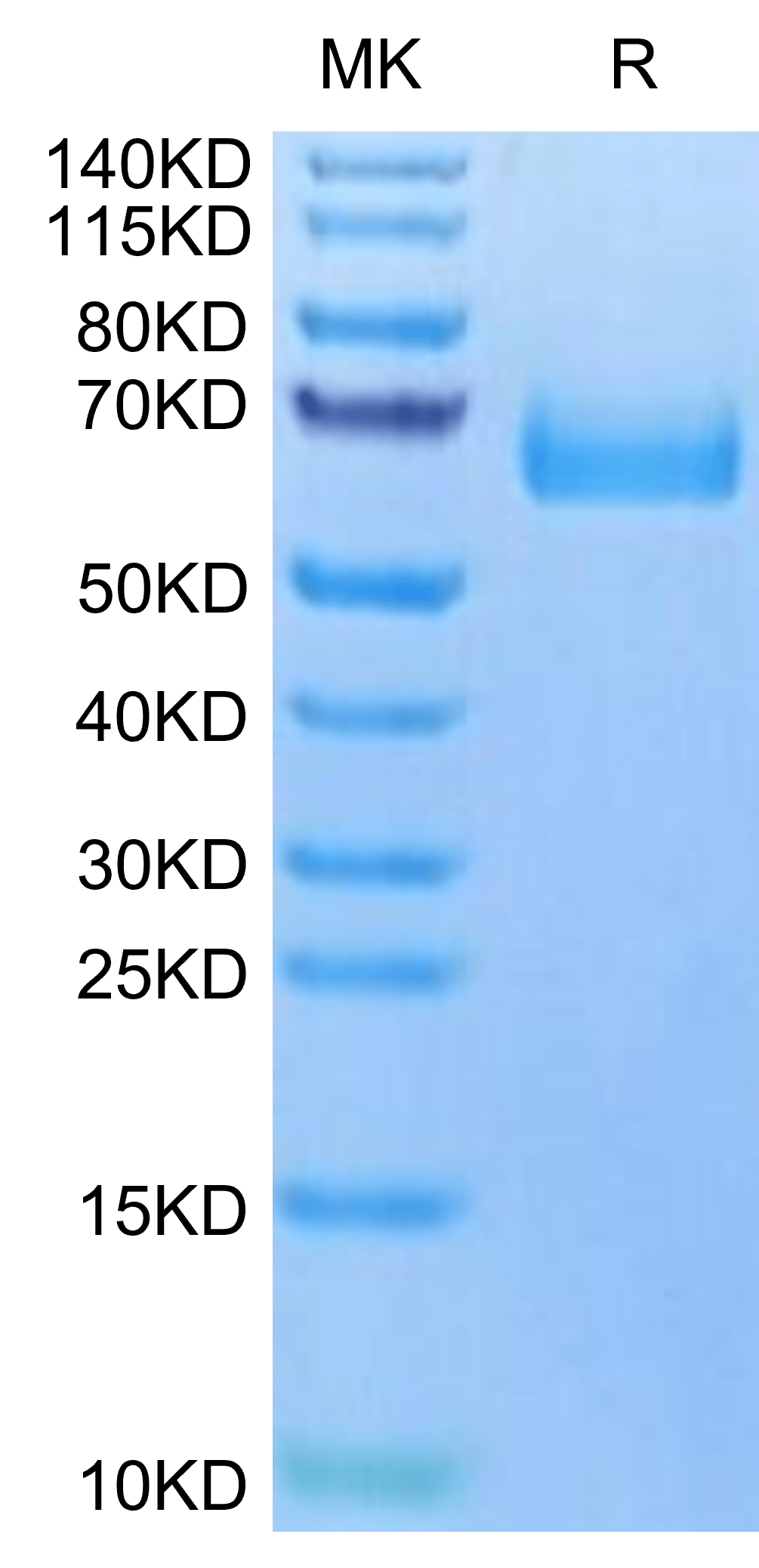
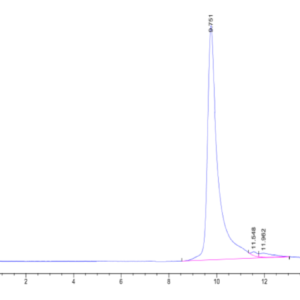
| molecular weight | The protein has a predicted MW of 32.69 kDa. Due to glycosylation, the protein migrates to 60-70 kDa based on Tris-Bis PAGE result. |
| available size | 100 µg, 500 µg |
| endotoxin | Less than 1EU per μg by the LAL method. |
Cynomolgus uPAR/PLAUR Protein 5081
$225.00 – $750.00
Summary
- Expression: HEK293
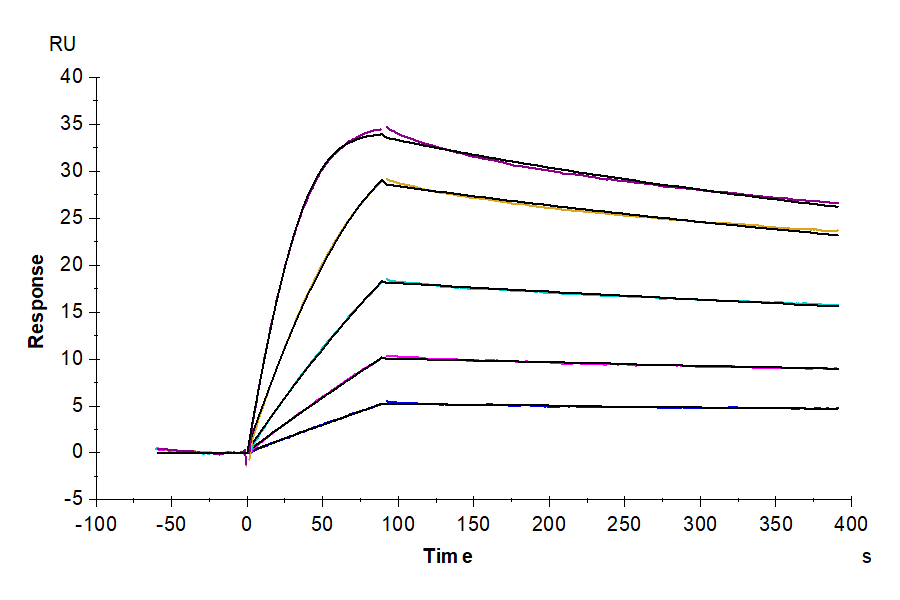
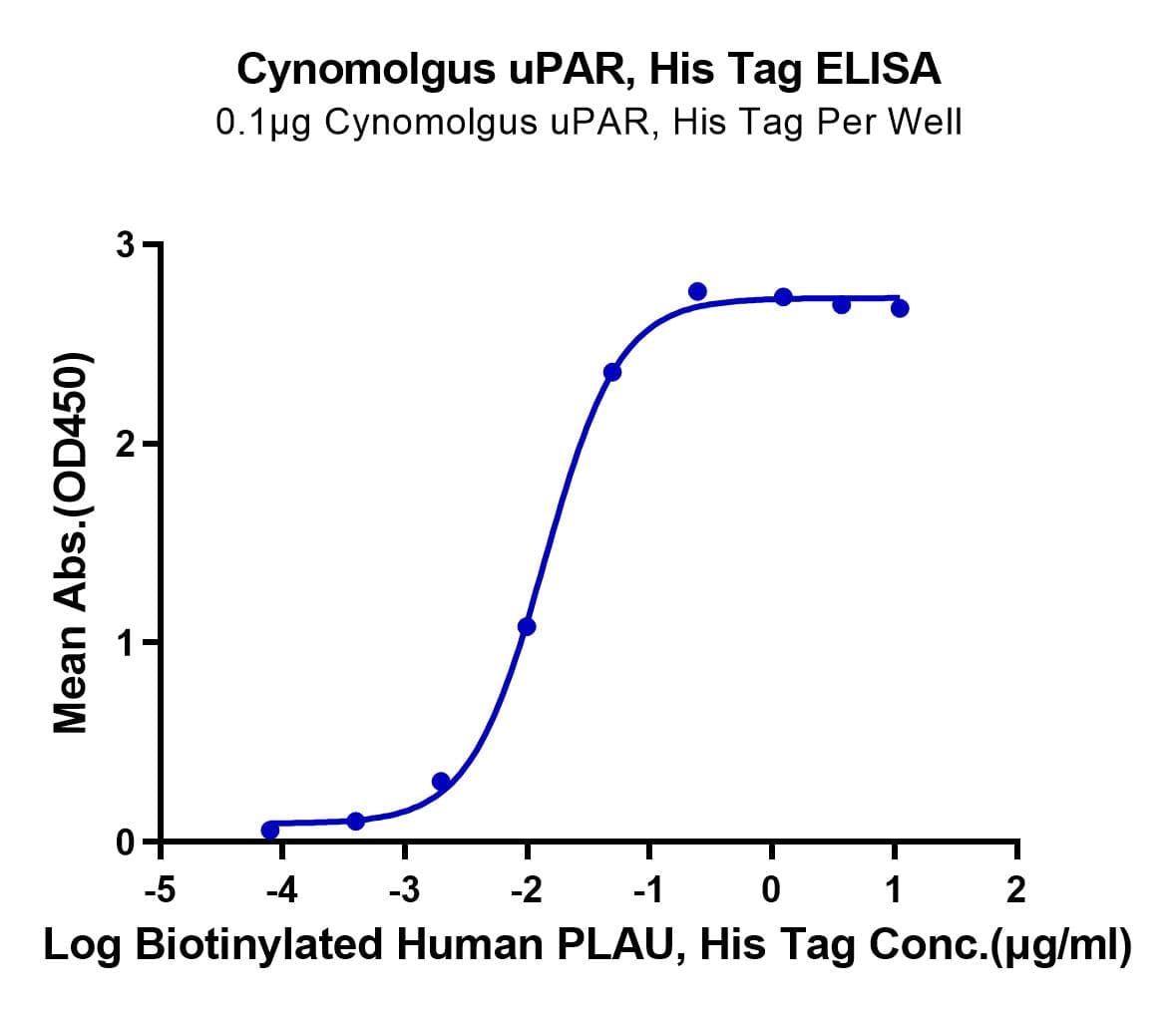
- Functional: Yes (ELISA)
- Amino Acid Range: Leu23-Gly305
Cynomolgus uPAR/PLAUR Protein 5081
| protein |
|---|
| Size and concentration 100, 500µg and lyophilized |
| Form Lyophilized |
| Storage Instructions Valid for 12 months from date of receipt when stored at -80°C. Recommend to aliquot the protein into smaller quantities for optimal storage. Please minimize freeze-thaw cycles. |
| Storage buffer Shipped at ambient temperature. |
| Purity > 95% as determined by Tris-Bis PAGE |
| target relevance |
|---|
| The receptor (u-PAR) for urokinase plasminogen activator (u-PA) is a three-domain protein, GPI-anchored to the cell surface, which focuses the enzymatic activity of u-PA, and allows the cell surface activation of plasminogen.Regulation of the activity of u-PA is also mediated by u-PAR. |
| Protein names Urokinase plasminogen activator surface receptor (U-PAR) (uPAR) (CD antigen CD87) |
| Gene names PLAUR,PLAUR UPAR |
| Mass 9541Da |
| Function Acts as a receptor for urokinase plasminogen activator. Plays a role in localizing and promoting plasmin formation. Mediates the proteolysis-independent signal transduction activation effects of U-PA. It is subject to negative-feedback regulation by U-PA which cleaves it into an inactive form (By similarity). |
| Subellular location Cell membrane. Cell projection, invadopodium membrane. Cell membrane ; Lipid-anchor, GPI-anchor. Note=Colocalized with FAP (seprase) preferentially at the cell surface of invadopodia membrane in a cytoskeleton-, integrin- and vitronectin-dependent manner. |
| Structure Monomer (Probable). Interacts (via the UPAR/Ly6 domains) with SRPX2. Interacts with MRC2. Interacts with FAP (seprase); the interaction occurs at the cell surface of invadopodia membrane. Interacts with SORL1 (via N-terminal ectodomain); this interaction decreases PLAUR internalization (By similarity). The ternary complex composed of PLAUR-PLAU-SERPINE1 also interacts with SORL1 (By similarity). |
| Target Relevance information above includes information from UniProt accession: Q9GK78 |
| The UniProt Consortium |
Data
Publications
Publications
| pmid | title | authors | citation |
|---|---|---|---|
| We haven't added any publications to our database yet. | |||
Protocols
| relevant to this product |
|---|
Documents
| # | ||
|---|---|---|
| Please enter your product and batch number here to retrieve product datasheet, SDS, and QC information. | ||